1TLD
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-TRYPSIN AT 1.5 ANGSTROMS RESOLUTION IN A CRYSTAL FORM WITH LOW MOLECULAR PACKING DENSITY. ACTIVE SITE GEOMETRY, ION PAIRS AND SOLVENT STRUCTURE | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, SULFATE ION | | Authors: | Bartunik, H.D, Summers, L.J, Bartsch, H.H. | | Deposit date: | 1989-07-24 | | Release date: | 1990-01-15 | | Last modified: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of bovine beta-trypsin at 1.5 A resolution in a crystal form with low molecular packing density. Active site geometry, ion pairs and solvent structure.
J.Mol.Biol., 210, 1989
|
|
1CZA
 
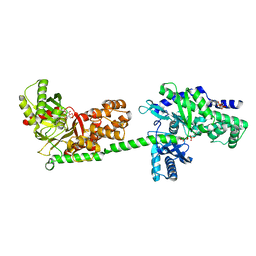 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I COMPLEXED WITH GLUCOSE, GLUCOSE-6-PHOSPHATE, AND ADP | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, ... | | Authors: | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
1DGK
 
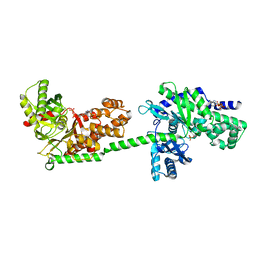 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I WITH GLUCOSE AND ADP IN THE ACTIVE SITE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, PHOSPHATE ION, ... | | Authors: | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
3K7M
 
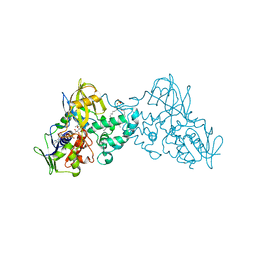 | | Crystal structure of 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3K7T
 
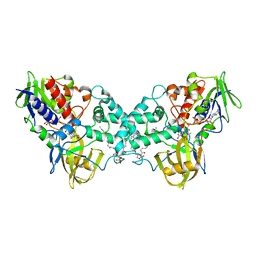 | | Crystal structure of apo-form 6-hydroxy-L-nicotine oxidase, crystal form P3121 | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3K7Q
 
 | | Crystal structure of substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, 6-hydroxy-L-nicotine oxidase, ... | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3SS9
 
 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.97 A resolution | | Descriptor: | D-serine dehydratase, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-08 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
3SS7
 
 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.55 A resolution | | Descriptor: | D-serine dehydratase, GLYCEROL, POTASSIUM ION, ... | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-01-18 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
2EWF
 
 | | Crystal structure of the site-specific DNA nickase N.BspD6I | | Descriptor: | BROMIDE ION, Nicking endonuclease N.BspD6I | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Perevyazova, T.A, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
3NG7
 
 | |
3NN0
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with nicotinamide | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-23 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK2
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with dopamine | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-18 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NGC
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with intermediate methylmyosmine product formed during catalytic turnover | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 5-(1-methyl-4,5-dihydro-1H-pyrrol-2-yl)pyridin-2-ol, 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-11 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NH3
 
 | |
3NHO
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with product bound at active site | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 1-(6-hydroxypyridin-3-yl)-4-(methylamino)butan-1-one, 6-hydroxy-L-nicotine oxidase, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK0
 
 | |
3NN6
 
 | | Crystal structure of inhibitor-bound in active centre 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, (2R,3S,4S)-5-({[(acetylcarbamoyl)amino]methyl}[(3S,4R)-6-amino-3,4-dimethylhexyl]amino)-2,3,4-trihydroxypentyl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 5-[(2R)-1-methylpyrrolidin-2-yl]pyridin-2-ol, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-23 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK1
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with serotonin | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-18 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
5LIQ
 
 | | The structure of C160S,C508S,C578S mutant of Nt.BspD6I nicking endonuclease at 0.185 nm resolution . | | Descriptor: | GLYCEROL, Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Artyukh, R.I, Perevyazova, T.A, Yunusova, A.K, Popov, A.N, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural features of Cysteine residues mutation of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
5LHC
 
 | | The structure of D456A mutant of Nt.BspD6I nicking endonuclease at 0.24 nm resolution . | | Descriptor: | GLYCEROL, Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Yunusova, A.K, Popov, A.N, Artyukh, R.I, Perevyazova, T.A, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-10 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural implication of activity loss by D456A mutant of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
5LIZ
 
 | | The structure of Nt.BspD6I nicking endonuclease with all cysteines mutated by serine residues at 0.19 nm resolution . | | Descriptor: | Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Artyukh, R.I, Perevyazova, T.A, Yunusova, A.K, Popov, A.N, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural features of Cysteine residues mutation of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
3CFF
 
 | |
3CFH
 
 | |
3CFA
 
 | |
1DO0
 
 | | ORTHORHOMBIC CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (HEAT SHOCK LOCUS U), ... | | Authors: | Bochtler, M, Hartmann, C, Song, H.K, Bourenkov, G.P, Bartunik, H.D. | | Deposit date: | 1999-12-18 | | Release date: | 2000-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of HsIU and the ATP-dependent protease HsIU-HsIV.
Nature, 403, 2000
|
|
