1BCT
 
 | |
6EVI
 
 | | solution NMR structure of EB1 C terminus (191-260) | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Barsukov, I.L, Almeida, T.B. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Targeting SxIP-EB1 interaction: An integrated approach to the discovery of small molecule modulators of dynamic binding sites.
Sci Rep, 7, 2017
|
|
6EVQ
 
 | |
2LNK
 
 | |
3IVF
 
 | | Crystal structure of the talin head FERM domain | | Descriptor: | Talin-1 | | Authors: | Elliott, P.R, Goult, B.T, Bate, N, Grossmann, J.G, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2009-09-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Structure of the talin head reveals a novel extended conformation of the FERM domain
Structure, 18, 2010
|
|
3PV1
 
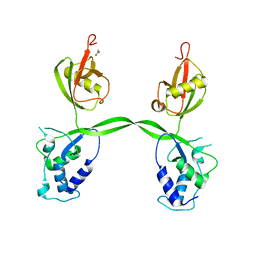 | | Crystal structure of the USP15 DUSP-UBL domains | | Descriptor: | ACETATE ION, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Elliott, P.R, Urbe, S, Clague, M.J, Barsukov, I.L. | | Deposit date: | 2010-12-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of the ubiquitin specific protease DUSP-UBL double domains.
Febs Lett., 585, 2011
|
|
4A3O
 
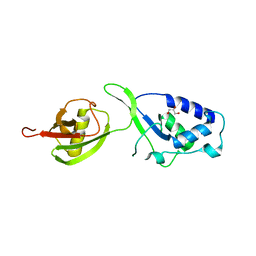 | | Crystal structure of the USP15 DUSP-UBL monomer | | Descriptor: | GLYCEROL, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
4A3P
 
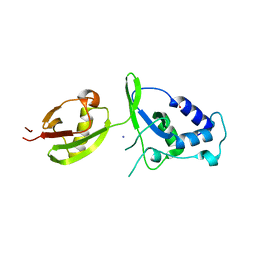 | | Structure of USP15 DUSP-UBL deletion mutant | | Descriptor: | ACETATE ION, IODIDE ION, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
7OLG
 
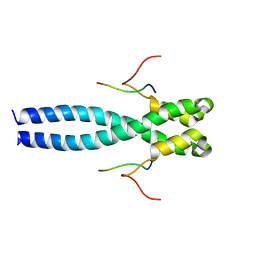 | | EB1 bound to MACF peptide | | Descriptor: | 11MACF, Microtubule-associated protein RP/EB family member 1 | | Authors: | Almeida, T.B, Barsukov, I.L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | EB1 bound to MACF peptide
To Be Published
|
|
3ZDL
 
 | | Vinculin head (1-258) in complex with a RIAM fragment | | Descriptor: | AMYLOID BETA A4 PRECURSOR PROTEIN-BINDING FAMILY B MEMBER 1-INTERACTING PROTEIN, VINCULIN | | Authors: | Zacharchenko, T, Elliott, P.R, Goult, B.T, Bate, N, Critchely, D.R, Barsukov, I.L. | | Deposit date: | 2012-11-28 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Riam and Vinculin Binding to Talin are Mutually Exclusive and Regulate Adhesion Assembly and Turnover.
J.Biol.Chem., 288, 2013
|
|
2QDQ
 
 | | Crystal structure of the talin dimerisation domain | | Descriptor: | Talin-1 | | Authors: | Gingras, A.R, Putz, N.S.M, Bate, N, Barsukov, I.L, Critchley, D.R.C. | | Deposit date: | 2007-06-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the C-terminal actin-binding domain of talin.
Embo J., 27, 2008
|
|
2X0C
 
 | | Crystal Structure of the R7R8 domains of Talin | | Descriptor: | TALIN-1 | | Authors: | Gingras, A.R, Goult, B.T, Bate, N, Barsukov, I.L, Emsley, J, Critchely, D.R. | | Deposit date: | 2009-12-08 | | Release date: | 2010-07-07 | | Last modified: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Central Region of Talin Has a Unique Fold that Binds Vinculin and Actin.
J.Biol.Chem., 285, 2010
|
|
1AO8
 
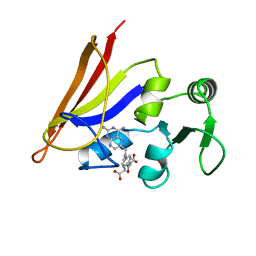 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE, NMR, 21 STRUCTURES | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE | | Authors: | Gargaro, A.R, Soteriou, A, Frenkiel, T.A, Bauer, C.J, Birdsall, B, Polshakov, V.I, Barsukov, I.L, Roberts, G.C.K, Feeney, J. | | Deposit date: | 1997-07-22 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex of Lactobacillus casei dihydrofolate reductase with methotrexate.
J.Mol.Biol., 277, 1998
|
|
5FZT
 
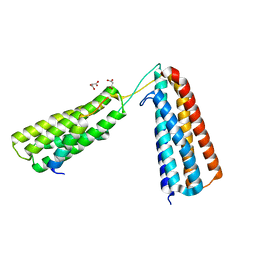 | | The crystal structure of R7R8 in complex with a DLC1 fragment. | | Descriptor: | MALONATE ION, RHO GTPASE-ACTIVATING PROTEIN 7, TALIN-1 | | Authors: | Zacharchenko, T, Qian, X, Goult, B.T, Jethwa, D, Almeida, T, Ballestrem, C, Critchley, D.R, Lowy, D.R, Barsukov, I.L. | | Deposit date: | 2016-03-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ld Motif Recognition by Talin: Structure of the Talin-Dlc1 Complex.
Structure, 24, 2016
|
|
3C1V
 
 | |
1ZVZ
 
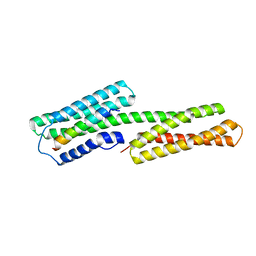 | | Vinculin Head (0-258) in Complex with the Talin Rod Residue 820-844 | | Descriptor: | Talin 1, Vinculin | | Authors: | Gingras, A.R, Ziegler, W.H, Barsukov, I.L, Roberts, G.C, Critchley, D.R, Emsley, J. | | Deposit date: | 2005-06-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod
J.Biol.Chem., 280, 2005
|
|
1ZW2
 
 | | Vinculin Head (0-258) in Complex with the Talin Rod residues 2345-2369 | | Descriptor: | Vinculin, talin | | Authors: | Gingras, A.R, Ziegler, W.H, Barsukov, I.L, Roberts, G.C, Critchley, D.R, Emsley, J. | | Deposit date: | 2005-06-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod
J.Biol.Chem., 280, 2005
|
|
2B0H
 
 | | Solution structure of VBS3 fragment of talin | | Descriptor: | Talin-1 | | Authors: | Gingras, A.R, Vogel, K.P, Steinhoff, H.J, Ziegler, W.H, Patel, B, Emsley, J, Critchley, D.R, Roberts, G.C, Barsukov, I.L. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Characterization of a Vinculin Binding Site in the Talin Rod
Biochemistry, 45, 2006
|
|
1U89
 
 | | Solution structure of VBS2 fragment of talin | | Descriptor: | Talin 1 | | Authors: | Fillingham, I, Gingras, A.R, Papagrigoriou, E, Patel, B, Emsley, J, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head.
Structure, 13, 2005
|
|
1U6H
 
 | | Vinculin head (0-258) in complex with the talin vinculin binding site 2 (849-879) | | Descriptor: | Talin, Vinculin | | Authors: | Fillingham, I, Gingras, A.R, Papagrigoriou, E, Patel, B, Emsley, J, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2004-07-30 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head.
Structure, 13, 2005
|
|
1XWJ
 
 | | Vinculin head (1-258) in complex with the talin vinculin binding site 3 (1945-1969) | | Descriptor: | Talin, Vinculin | | Authors: | Gingras, A.R, Papagrigoriou, E, Barsukov, I.L, Critchley, D.R, Emsley, J. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Dynamic Characterization of a Vinculin Binding Site in the Talin Rod
Biochemistry, 45, 2006
|
|
1ZW3
 
 | | Vinculin Head (0-258) in Complex with the Talin Rod residues 1630-1652 | | Descriptor: | Talin 1, Vinculin | | Authors: | Gingras, A.R, Ziegler, W.H, Barsukov, I.L, Roberts, G.C, Critchley, D.R, Emsley, J. | | Deposit date: | 2005-06-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod
J.Biol.Chem., 280, 2005
|
|
1GRM
 
 | | REFINEMENT OF THE SPATIAL STRUCTURE OF THE GRAMICIDIN A TRANSMEMBRANE ION-CHANNEL (RUSSIAN) | | Descriptor: | GRAMICIDIN A | | Authors: | Arseniev, A.S, Barsukov, I.L, Lomize, A.L, Orekhov, V.Y, Bystrov, V.F. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Refinement of the Spatial Structure of the Gramicidin a Ion Channel
Biol.Membr.(Ussr), 18, 1992
|
|
1SJ8
 
 | | Solution Structure of the R1R2 Domains of Talin | | Descriptor: | Talin 1 | | Authors: | Papagrigoriou, E, Gingras, A.R, Barsukov, I.L, Critchley, D.R, Emsley, J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle
EMBO J., 23, 2004
|
|
1SJ7
 
 | | Crystal Structure of Talin Rod 482-655 | | Descriptor: | PLATINUM (II) ION, Talin 1 | | Authors: | Papagrigoriou, E, Gingras, A.R, Barsukov, I.L, Critchley, D.R, Emsley, J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle
EMBO J., 23, 2004
|
|
