6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
1KWI
 
 | | Crystal Structure Analysis of the Cathelicidin Motif of Protegrins | | Descriptor: | Protegrin-3 Precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
2B68
 
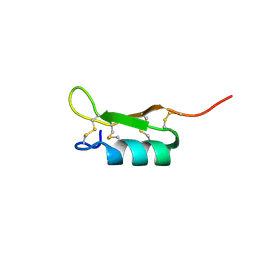 | | Solution structure of the recombinant Crassostrea gigas defensin | | Descriptor: | defensin | | Authors: | Gueguen, Y, Amaury, H, Aumelas, A, Garnier, J, Fievet, J, Escoubas, J.M, Bulet, P, Gonzales, M, Lelong, C, Favrel, P, Bachere, E. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-29 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Defensin from the Oyster Crassostrea gigas: recombinant production, folding, solution structure, antimicrobial activities and gene expression
J.Biol.Chem., 281, 2006
|
|
2JOB
 
 | | Solution structure of an antilipopolysaccharide factor from shrimp and its possible Lipid A binding site | | Descriptor: | antilipopolysaccharide factor | | Authors: | Yang, Y, Boze, H, Chemardin, P, Padilla, A, Moulin, G, Tassanakajon, A, Pugniere, M, Roquet, F, Gueguen, Y, Bachere, E, Aumelas, A. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: Model of the possible lipid A-binding site
Biopolymers, 91, 2009
|
|
1PAE
 
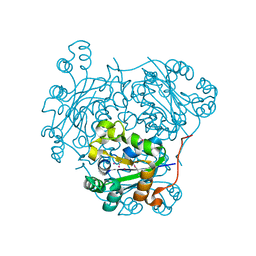 | | nucleoside diphosphate kinase | | Descriptor: | Nucleoside diphosphate kinase, cytosolic, SELENIUM ATOM | | Authors: | Strub, M.-P, Hoh, F, Sanchez, J.-F, Strub, J.M, Bock, A, Aumelas, A, Dumas, C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-11-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing
Structure, 11, 2003
|
|
1PFP
 
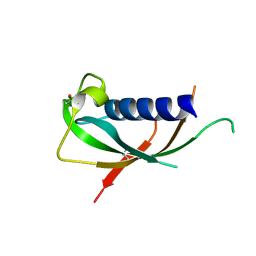 | | CATHELIN-LIKE MOTIF OF PROTEGRIN-3 | | Descriptor: | Protegrin 3 | | Authors: | Strub, M.-P, Hoh, F, Sanchez, J.-F, Strub, J.M, Bock, A, Aumelas, A, Dumas, C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing
Structure, 11, 2003
|
|
1XV3
 
 | | NMR structure of the synthetic penaeidin 4 | | Descriptor: | Penaeidin-4d | | Authors: | Cuthbertson, B.J, Yang, Y, Bullesbach, E.E, Bachere, E, Gross, P.S, Aumelas, A. | | Deposit date: | 2004-10-27 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of synthetic penaeidin-4 with structural and functional comparisons to penaeidin-3
J.Biol.Chem., 280, 2005
|
|
1T7H
 
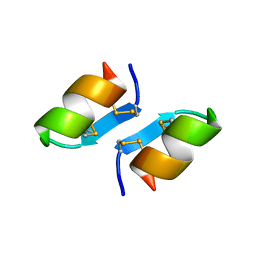 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | Descriptor: | Endothelin-1 | | Authors: | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | Deposit date: | 2004-05-10 | | Release date: | 2004-12-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
1UEO
 
 | | Solution structure of the [T8A]-Penaeidin-3 | | Descriptor: | Penaeidin-3a | | Authors: | Yang, Y, Poncet, J, Garnier, J, Zatylny, C, Bachere, E, Aumelas, A. | | Deposit date: | 2003-05-20 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide
J.Biol.Chem., 278, 2003
|
|
2EEM
 
 | |
2JOS
 
 | |
2L24
 
 | | Antimicrobial peptide | | Descriptor: | Hemagglutinin | | Authors: | Zhu, S, Aumelas, A, Gao, B. | | Deposit date: | 2010-08-10 | | Release date: | 2011-06-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Convergent evolution-guided design of antimicrobial peptides derived from influenza A virus hemagglutinin.
J.Med.Chem., 54, 2011
|
|
1LXE
 
 | | CRYSTAL STRUCTURE OF THE CATHELICIDIN MOTIF OF PROTEGRINS | | Descriptor: | protegrin-3 precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-06-05 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
1N5H
 
 | | Solution structure of the cathelin-like domain of protegrins (the R87-P88 and D118-P119 amide bonds are in the cis conformation) | | Descriptor: | protegrins | | Authors: | Yang, Y, Sanchez, J.F, Strub, M.P, Brutscher, B, Aumelas, A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Cathelin-like domain of the protegrin-3 Precursor
Biochemistry, 42, 2003
|
|
1N5P
 
 | | Solution structure of the cathelin-like domain of protegrins (all amide bonds involving proline residues are in trans conformation) | | Descriptor: | protegrins | | Authors: | Yang, Y, Sanchez, J.F, Strub, M.P, Brutscher, B, Aumelas, A. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Cathelin-like domain of the protegrin-3 Precursor
Biochemistry, 42, 2003
|
|
1FJN
 
 | |
2GFI
 
 | | Crystal structure of the phytase from D. castellii at 2.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Hoh, F. | | Deposit date: | 2006-03-22 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Debaryomyces castellii CBS 2923 phytase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
