4WT8
 
 | | Crystal Structure of bactobolin A bound to 70S ribosome-tRNA complex | | Descriptor: | 23S rRNA (2899-MER), 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Amunts, A, Fiedorczuk, K, Ramakrishnan, V. | | Deposit date: | 2014-10-29 | | Release date: | 2015-01-21 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bactobolin A Binds to a Site on the 70S Ribosome Distinct from Previously Seen Antibiotics.
J.Mol.Biol., 427, 2015
|
|
2O01
 
 | | The Structure of a plant photosystem I supercomplex at 3.4 Angstrom resolution | | Descriptor: | AT3g54890, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Amunts, A, Drory, O, Nelson, N. | | Deposit date: | 2006-11-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structure of a plant photosystem I supercomplex at 3.4 A resolution.
Nature, 447, 2007
|
|
2WSC
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-04 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure Determination and Improved Model of Plant Photosystem I.
J.Biol.Chem., 285, 2010
|
|
2WSE
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-05 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure determination and improved model of plant photosystem I
J. Biol. Chem., 285, 2010
|
|
2WSF
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-05 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structure determination and improved model of plant photosystem I.
J. Biol. Chem., 285, 2010
|
|
3J9M
 
 | | Structure of the human mitochondrial ribosome (class 1) | | Descriptor: | 12S rRNA, 16S rRNA, E-site tRNA, ... | | Authors: | Amunts, A, Brown, A, Toots, J, Scheres, S.H, Ramakrishnan, V. | | Deposit date: | 2015-02-08 | | Release date: | 2015-04-15 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome. The structure of the human mitochondrial ribosome.
Science, 348, 2015
|
|
3J6B
 
 | | Structure of the yeast mitochondrial large ribosomal subunit | | Descriptor: | 21S ribosomal RNA, 54S ribosomal protein IMG1, mitochondrial, ... | | Authors: | Amunts, A, Brown, A, Bai, X.C, Llacer, J.L, Hussain, T, Emsley, P, Long, F, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the yeast mitochondrial large ribosomal subunit.
Science, 343, 2014
|
|
6YWS
 
 | | The structure of the large subunit of the mitoribosome from Neurospora crassa | | Descriptor: | 50S ribosomal protein L14, 50S ribosomal protein L17, 50S ribosomal protein L24, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWX
 
 | | The structure of the mitoribosome from Neurospora crassa with tRNA bound to the E-site | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWY
 
 | | The structure of the mitoribosome from Neurospora crassa with bound tRNA at the P-site | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWE
 
 | | The structure of the mitoribosome from Neurospora crassa in the P/E tRNA bound state | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YW5
 
 | | The structure of the small subunit of the mitoribosome from Neurospora crassa | | Descriptor: | 16S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWV
 
 | | The structure of the Atp25 bound assembly intermediate of the mitoribosome from Neurospora crassa | | Descriptor: | 23 S rRNA, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
7P2E
 
 | | Human mitochondrial ribosome small subunit in complex with IF3, GMPPMP and streptomycin | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Singh, V, Rorbach, J, Amunts, A. | | Deposit date: | 2021-07-05 | | Release date: | 2022-07-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the mitoribosomal small subunit with streptomycin reveals Fe-S clusters and physiological molecules.
Elife, 11, 2022
|
|
7AOI
 
 | | Trypanosoma brucei mitochondrial ribosome large subunit assembly intermediate | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Tobiasson, V, Gahura, O, Aibara, S, Baradaran, R, Zikova, A, Amunts, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit.
Embo J., 40, 2021
|
|
8OM2
 
 | | Small subunit of yeast mitochondrial ribosome in complex with METTL17/Rsm22. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8OM4
 
 | | Small subunit of yeast mitochondrial ribosome. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8OM3
 
 | | Small subunit of yeast mitochondrial ribosome in complex with IF3/Aim23. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8B6F
 
 | | Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6H
 
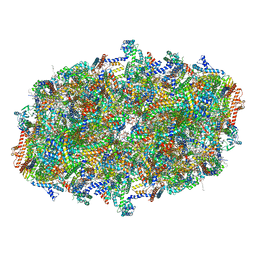 | | Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6J
 
 | | Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apocytochrome b, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6G
 
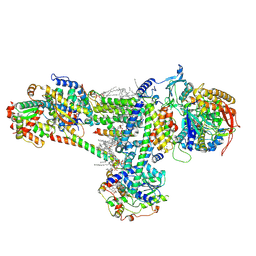 | | Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
7OG4
 
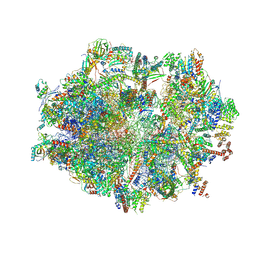 | | Human mitochondrial ribosome in complex with P/E-tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2021-05-06 | | Release date: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
8BQS
 
 | | Cryo-EM structure of the I-II-III2-IV2 respiratory supercomplex from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mitochondrial membrane bending by I-II-III2-IV2 supercomplex
To Be Published
|
|
8BCV
 
 | | Photosystem I assembly intermediate of Avena sativa | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A, Nelson, N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Photosystem I assembly intermediate of Avena sativa
To Be Published
|
|
