1ESY
 
 | |
1F6U
 
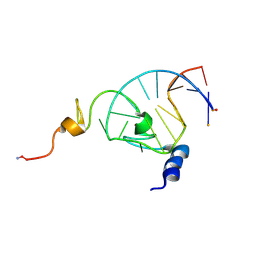 | | NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop sl2 of the psi-RNA packaging signal. Implications for genome recognition | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, HIV-1 STEM-LOOP SL2 FROM PSI-RNA PACKAGING, ZINC ION | | Authors: | Amarasinghe, G.K, De Guzman, R.N, Turner, R.B, Chancellor, K.J, Summers, M.F. | | Deposit date: | 2000-06-23 | | Release date: | 2000-10-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition.
J.Mol.Biol., 301, 2000
|
|
3FKE
 
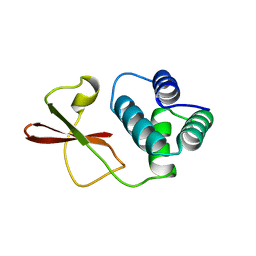 | | Structure of the Ebola VP35 Interferon Inhibitory Domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Amarasinghe, G.K, Leung, D.W, Ginder, N.D, Honzatko, R.B, Nix, J, Basler, C.F, Fulton, D.B. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Ebola VP35 interferon inhibitory domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6C6K
 
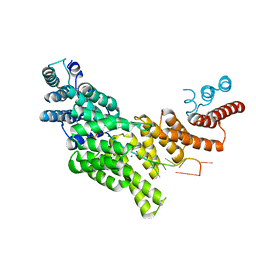 | | Structural basis for preferential recognition of cap 0 RNA by a human IFIT1-IFIT3 protein complex | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, Interferon-induced protein with tetratricopeptide repeats 3, MAGNESIUM ION, ... | | Authors: | Amarasinghe, G.K, Leung, D.W, Johnson, B, Xu, W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability.
Immunity, 48, 2018
|
|
4YPI
 
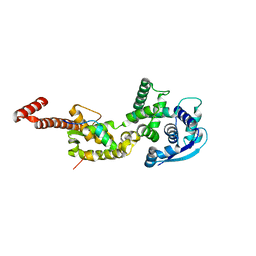 | | Structure of Ebola virus nucleoprotein N-terminal fragment bound to a peptide derived from Ebola VP35 | | Descriptor: | Nucleoprotein, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Borek, D.M, Binning, J.M, Otwinowski, Z, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.
Cell Rep, 11, 2015
|
|
6NTV
 
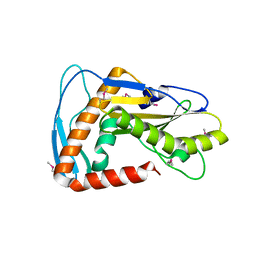 | | SFTSV L endonuclease domain | | Descriptor: | RNA polymerase | | Authors: | Wang, W, Amarasinghe, G.K. | | Deposit date: | 2019-01-30 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cap-Snatching SFTSV Endonuclease Domain Is an Antiviral Target.
Cell Rep, 30, 2020
|
|
6C54
 
 | | Ebola nucleoprotein nucleocapsid-like assembly and the asymmetric unit | | Descriptor: | Nucleoprotein | | Authors: | Su, Z, Wu, C, Pintilie, G.D, Chiu, W, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2018-01-13 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Electron Cryo-microscopy Structure of Ebola Virus Nucleoprotein Reveals a Mechanism for Nucleocapsid-like Assembly.
Cell, 172, 2018
|
|
8D3F
 
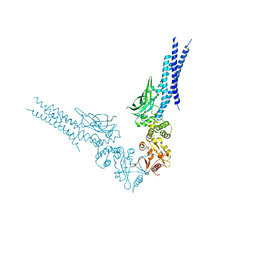 | | Crystal structure of human STAT1 in complex with the repeat region from Toxoplasma protein TgIST | | Descriptor: | Signal transducer and activator of transcription 1-alpha/beta,Inhibitor of STAT1-dependent transcription TgIST | | Authors: | Huang, Z, Liu, H, Nix, J.C, Amarasinghe, G.K, Sibley, L.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The intrinsically disordered protein TgIST from Toxoplasma gondii inhibits STAT1 signaling by blocking cofactor recruitment.
Nat Commun, 13, 2022
|
|
4M0Q
 
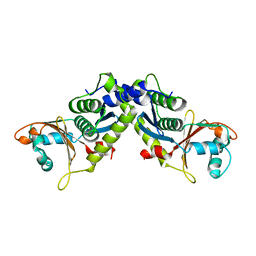 | | Ebola virus VP24 structure | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Xu, W, Leung, D.W, Borek, D, Amarasinghe, G.K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | The Marburg Virus VP24 Protein Interacts with Keap1 to Activate the Cytoprotective Antioxidant Response Pathway.
Cell Rep, 6, 2014
|
|
5VAP
 
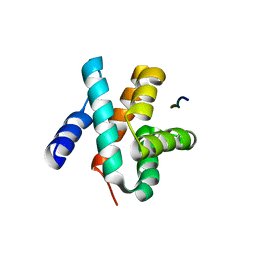 | | Crystal structure of eVP30 C-terminus and eNP peptide | | Descriptor: | Minor nucleoprotein VP30, NP | | Authors: | XU, W, WU, C, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis.
Nat Commun, 8, 2017
|
|
5VAO
 
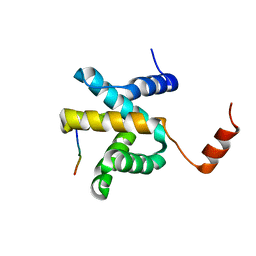 | | Crystal structure of eVP30 C-terminus and eNP peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | XU, W, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis.
Nat Commun, 8, 2017
|
|
5VJ2
 
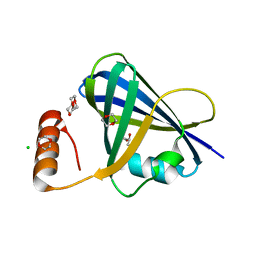 | | Structure of human respiratory syncytial virus non-structural protein 1 (NS1) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Leung, D.W, Borek, D.M, Amarasinghe, G.K. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for human respiratory syncytial virus NS1-mediated modulation of host responses.
Nat Microbiol, 2, 2017
|
|
3L25
 
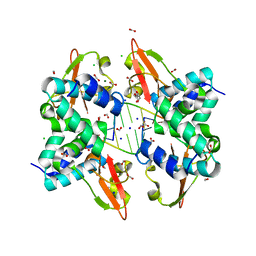 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L28
 
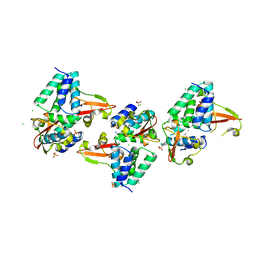 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain K339A mutant | | Descriptor: | CHLORIDE ION, Polymerase cofactor VP35, SODIUM ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L29
 
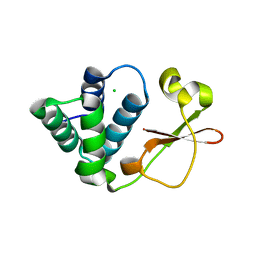 | | Crystal Structure of Zaire Ebola VP35 interferon inhibitory domain K319A/R322A mutant | | Descriptor: | CHLORIDE ION, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Ramanan, P, Borek, D.M, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs.
J.Virol., 84, 2010
|
|
3L26
 
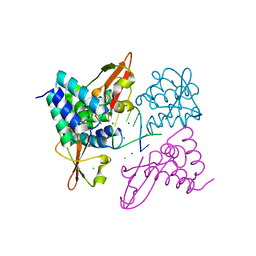 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L27
 
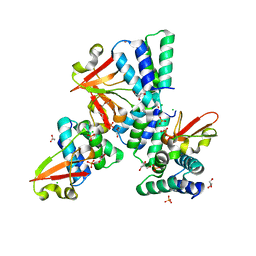 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain R312A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6DKU
 
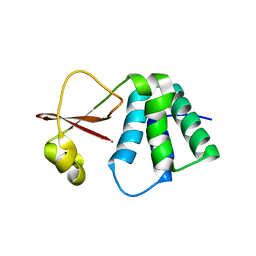 | | Crystal structure of Myotis VP35 mutant of interferon inhibitory domain | | Descriptor: | VP35 | | Authors: | Liu, H, Ginell, G.M, Keefe, L.J, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conservation of Structure and Immune Antagonist Functions of Filoviral VP35 Homologs Present in Microbat Genomes.
Cell Rep, 24, 2018
|
|
6E5X
 
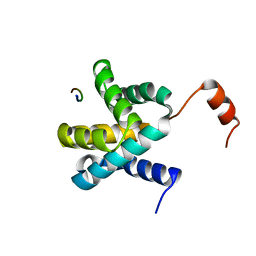 | | Crystal structure of Ebola virus VP30 C-terminus/RBBP6 peptide complex | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase RBBP6, Minor nucleoprotein VP30 | | Authors: | Liu, D, Small, G.I, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-07-23 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication.
Cell, 175, 2018
|
|
3L2A
 
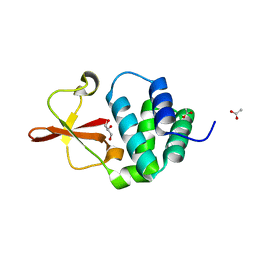 | | Crystal structure of Reston Ebola VP35 interferon inhibitory domain | | Descriptor: | ACETIC ACID, GLYCEROL, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Farahbakhsh, M, Borek, D.M, Prins, K.C, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Characterization of Reston Ebola Virus VP35 Interferon Inhibitory Domain.
J.Mol.Biol., 399, 2010
|
|
7LDK
 
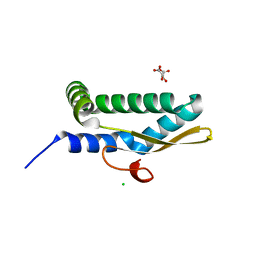 | | Structure of human respiratory syncytial virus nonstructural protein 2 (NS2) | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, Non-structural protein 2 | | Authors: | Chatterjee, S, Borek, D, Otwinowski, Z, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2021-01-13 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for IFN antagonism by human respiratory syncytial virus nonstructural protein 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4GHL
 
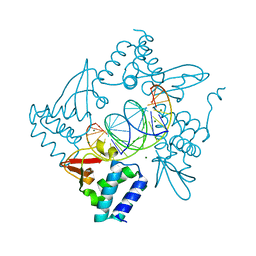 | | Structural Basis for Marburg virus VP35 mediate immune evasion mechanisms | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ramanan, P, Borek, D.M, Otwinowski, Z, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for Marburg virus VP35-mediated immune evasion mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4IBG
 
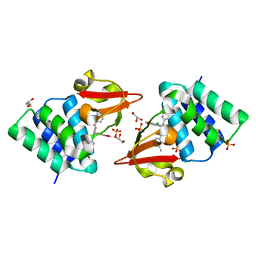 | | Ebola virus VP35 bound to small molecule | | Descriptor: | GLYCEROL, PHOSPHATE ION, Polymerase cofactor VP35, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBB
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | Polymerase cofactor VP35, {4-[(5R)-3-hydroxy-2-oxo-4-(thiophen-2-ylcarbonyl)-5-(2,4,5-trimethylphenyl)-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBD
 
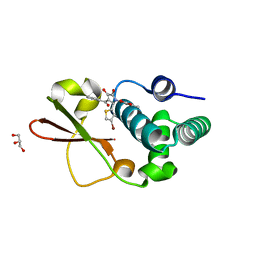 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-methylbenzoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
