3IEF
 
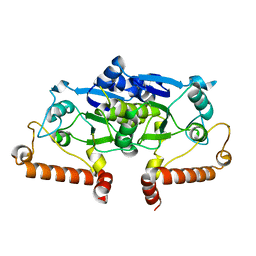 | |
3CXK
 
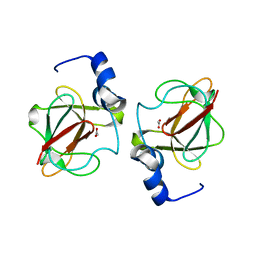 | | 1.7 A Crystal structure of methionine-R-sulfoxide reductase from Burkholderia pseudomallei: crystallization in a microfluidic crystal card. | | Descriptor: | ACETATE ION, Methionine-R-sulfoxide reductase, ZINC ION | | Authors: | Lovell, S, Gerdts, C, Staker, B, Craigen, D, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The plug-based nanovolume Microcapillary Protein Crystallization System (MPCS).
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3D4S
 
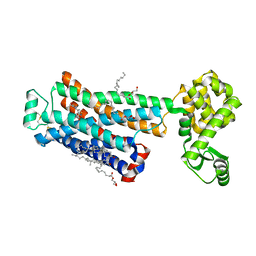 | | Cholesterol bound form of human beta2 adrenergic receptor. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, Beta-2 adrenergic receptor/T4-lysozyme chimera, ... | | Authors: | Hanson, M.A, Cherezov, V, Roth, C.B, Griffith, M.T, Jaakola, V.-P, Chien, E.Y.T, Velasquez, J, Kuhn, P, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor.
Structure, 16, 2008
|
|
3D38
 
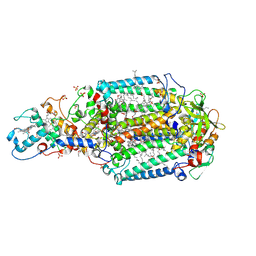 | | Crystal structure of new trigonal form of photosynthetic reaction center from Blastochloris viridis. Crystals grown in microfluidics by detergent capture. | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Nachtergaele, S.H.M, Seddon, A.M, Tereshko, V, Ponomarenko, N, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Simple host-guest chemistry to modulate the process of concentration and crystallization of membrane proteins by detergent capture in a microfluidic device.
J.Am.Chem.Soc., 130, 2008
|
|
2RHJ
 
 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ with Two Sulfate Ions and Sodium Ion in the Nucleotide Pocket | | Descriptor: | ACETATE ION, Cell Division Protein ftsZ, SODIUM ION, ... | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
2RH1
 
 | | High resolution crystal structure of human B2-adrenergic G protein-coupled receptor. | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Cherezov, V, Rosenbaum, D.M, Hanson, M.A, Rasmussen, S.G.F, Thian, F.S, Kobilka, T.S, Choi, H.J, Kuhn, P, Weis, W.I, Kobilka, B.K, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2007-10-05 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor.
Science, 318, 2007
|
|
2RHO
 
 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ NCS Dimer with Bound GDP and GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell Division Protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
2RHH
 
 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ with Bound Sulfate Ion | | Descriptor: | Cell Division Protein ftsZ, SULFATE ION | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
2RHL
 
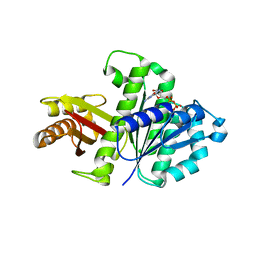 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ NCS Dimer with Bound GDP | | Descriptor: | Cell Division Protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
3BJI
 
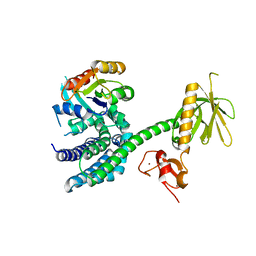 | | Structural Basis of Promiscuous Guanine Nucleotide Exchange by the T-Cell Essential Vav1 | | Descriptor: | Proto-oncogene vav, Ras-related C3 botulinum toxin substrate 1 precursor, ZINC ION | | Authors: | Chrencik, J.E, Brooun, A, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-04 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of guanine nucleotide exchange mediated by the T-cell essential Vav1.
J.Mol.Biol., 380, 2008
|
|
3II9
 
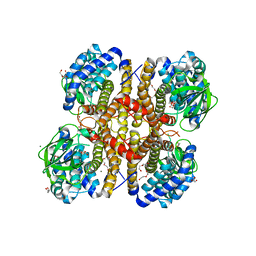 | | Crystal structure of glutaryl-coa dehydrogenase from Burkholderia pseudomallei at 1.73 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glutaryl-CoA dehydrogenase, ... | | Authors: | Ismagilov, R.F, Li, L, Du, W.B, Staker, B, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | User-loaded SlipChip for equipment-free multiplexed nanoliter-scale experiments.
J.Am.Chem.Soc., 132, 2010
|
|
2BBA
 
 | |
3ODU
 
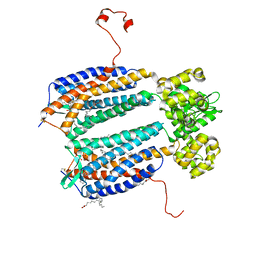 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3KJR
 
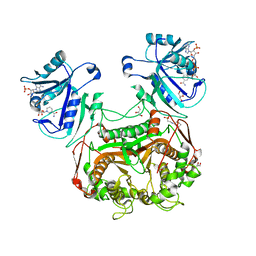 | | Crystal structure of dihydrofolate reductase/thymidylate synthase from Babesia bovis determined using SlipChip based microfluidics | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dihydrofolate reductase/thymidylate synthase, GLYCEROL, ... | | Authors: | Li, L, Du, W, Edwards, T.E, Staker, B.L, Phan, I, Stacy, R, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiparameter screening on SlipChip used for nanoliter protein crystallization combining free interface diffusion and microbatch methods.
J.Am.Chem.Soc., 132, 2010
|
|
3OE6
 
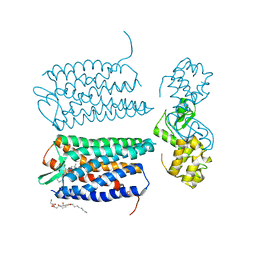 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3EML
 
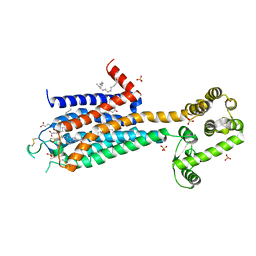 | | The 2.6 A Crystal Structure of a Human A2A Adenosine Receptor bound to ZM241385. | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Human Adenosine A2A receptor/T4 lysozyme chimera, STEARIC ACID, ... | | Authors: | Jaakola, V.-P, Griffith, M.T, Hanson, M.A, Cherezov, V, Chien, E.Y.T, Lane, J.R, Ijzerman, A.P, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist.
Science, 322, 2008
|
|
3BLA
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG153249 | | Descriptor: | 5-[(1S)-1-(3-chlorophenyl)ethoxy]quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
3BL9
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG157493 | | Descriptor: | 5-{[1-(2,3-dichlorobenzyl)piperidin-4-yl]methoxy}quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
3BL7
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG156844 | | Descriptor: | 5-{[1-(2-fluorobenzyl)piperidin-4-yl]methoxy}quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
3NY8
 
 | | Crystal structure of the human beta2 adrenergic receptor in complex with the inverse agonist ICI 118,551 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3S)-1-[(7-methyl-2,3-dihydro-1H-inden-4-yl)oxy]-3-[(1-methylethyl)amino]butan-2-ol, Beta-2 adrenergic receptor, ... | | Authors: | Wacker, D, Fenalti, G, Brown, M.A, Katritch, V, Abagyan, R, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography
J.Am.Chem.Soc., 132, 2010
|
|
3OE9
 
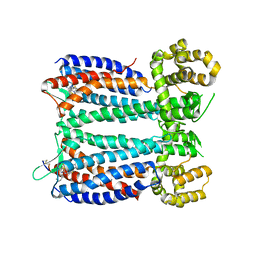 | | Crystal structure of the chemokine CXCR4 receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE8
 
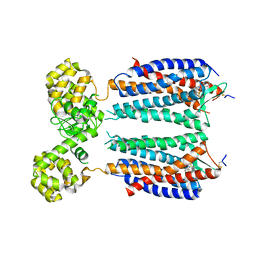 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3NY9
 
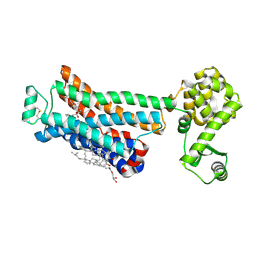 | | Crystal structure of the human beta2 adrenergic receptor in complex with a novel inverse agonist | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Fenalti, G, Wacker, D, Brown, M.A, Katritch, V, Abagyan, R, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography
J.Am.Chem.Soc., 132, 2010
|
|
3OE0
 
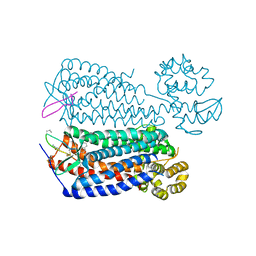 | | Crystal structure of the CXCR4 chemokine receptor in complex with a cyclic peptide antagonist CVX15 | | Descriptor: | C-X-C chemokine receptor type 4, Lysozyme Chimera, Polyphemusin analog, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3IFT
 
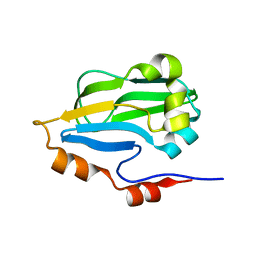 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
