5K9B
 
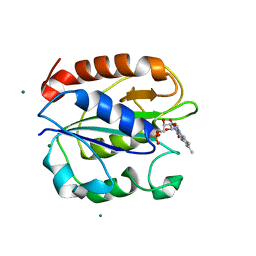 | | Azotobacter vinelandii Flavodoxin II | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin-2, MAGNESIUM ION | | Authors: | Rees, D.C, Segal, H.M, Spatzal, T. | | Deposit date: | 2016-05-31 | | Release date: | 2017-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.174 Å) | | Cite: | Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II.
Protein Sci., 26, 2017
|
|
4CPA
 
 | |
407D
 
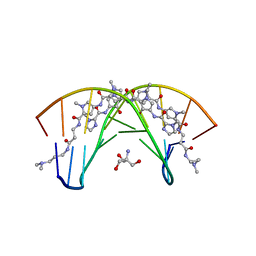 | | STRUCTURAL BASIS FOR RECOGNITION OF A-T AND T-A BASE PAIRS IN THE MINOR GROOVE OF B-DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*G)-3'), ~{N}-[5-[[5-[[5-[[3-[3-(dimethylamino)propylamino]-3-oxidanylidene-propyl]carbamoyl]-1-methyl-pyrrol-3-yl]carbamoyl]-1-methyl-pyrrol-3-yl]carbamoyl]-1-methyl-4-oxidanyl-pyrrol-3-yl]-1-methyl-imidazole-2-carboxamide | | Authors: | Rees, D.C. | | Deposit date: | 1998-06-24 | | Release date: | 1998-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA.
Science, 282, 1998
|
|
2OAU
 
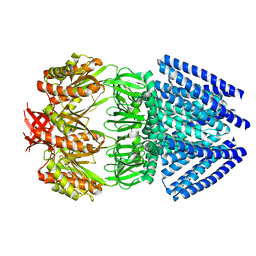 | | Mechanosensitive Channel of Small Conductance (MscS) | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Rees, D.C, Bass, R.B, Steinbacher, S, Strop, P, Barclay, M.T. | | Deposit date: | 2006-12-17 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of the Prokaryotic Mechanosensitive Channels MscL and MscS
CURRENT TOPICS IN MEMBRANES, 58, 2007
|
|
2OAR
 
 | | Mechanosensitive Channel of Large Conductance (MscL) | | Descriptor: | GOLD ION, Large-conductance mechanosensitive channel | | Authors: | Rees, D.C, Chang, G, Spencer, R.H, Lee, A.T, Steinbacher, S, Strop, P. | | Deposit date: | 2006-12-17 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of the Prokaryotic Mechanosensitive Channels MscL and MscS
Current Topics in Membranes, 58, 2007
|
|
3DHW
 
 | | Crystal structure of methionine importer MetNI | | Descriptor: | D-methionine transport system permease protein metI, Methionine import ATP-binding protein metN | | Authors: | Rees, D.C, Kaiser, J.T, Kadaba, N.S, Johnson, E, Lee, A.T. | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation.
Science, 321, 2008
|
|
2MIN
 
 | | NITROGENASE MOFE PROTEIN FROM AZOTOBACTER VINELANDII, OXIDIZED STATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Peters, J.W, Stowell, M.H.B, Soltis, S.M, Day, M.W, Kim, J, Rees, D.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-dependent structural changes in the nitrogenase P-cluster.
Biochemistry, 36, 1997
|
|
4WZB
 
 | | Crystal Structure of MgAMPPCP-bound Av2-Av1 complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-11-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nitrogenase complexes: multiple docking sites for a nucleotide switch protein.
Science, 309, 2005
|
|
1AIG
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE D+QB-CHARGE SEPARATED STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stowell, M.H.B, Mcphillips, T.M, Soltis, S.M, Rees, D.C, Abresch, E, Feher, G. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer.
Science, 276, 1997
|
|
1AIJ
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE CHARGE-NEUTRAL DQAQB STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stowell, M.H.B, Mcphillips, T.M, Soltis, S.M, Rees, D.C, Abresch, E, Feher, G. | | Deposit date: | 1997-04-18 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer.
Science, 276, 1997
|
|
1WOD
 
 | | CRYSTAL STRUCTURE OF MODA, A MOLYBDATE PROTEIN, COMPLEXED WITH TUNGSTATE | | Descriptor: | MODA, TUNGSTATE(VI)ION | | Authors: | Hu, Y, Rech, S, Gunsalus, R.P, Rees, D.C. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdate binding protein ModA.
Nat.Struct.Biol., 4, 1997
|
|
1PST
 
 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
1PSS
 
 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
4WES
 
 | | Nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 1.08 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, ... | | Authors: | Zhang, L.M, Morrison, C.N, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08 angstrom resolution: comparison with the Azotobacter vinelandii MoFe protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WN9
 
 | | Structure of the Nitrogenase MoFe Protein from Clostridium pasteurianum Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
4WNA
 
 | | Structure of the Nitrogenase MoFe Protein from Azotobacter vinelandii Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Zhang, L, Einsle, O, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
4WZA
 
 | | Asymmetric Nucleotide Binding in the Nitrogenase Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-11-19 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8995 Å) | | Cite: | Structural evidence for asymmetrical nucleotide interactions in nitrogenase.
J.Am.Chem.Soc., 137, 2015
|
|
4RCR
 
 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
3MIN
 
 | | NITROGENASE MOFE PROTEIN FROM AZOTOBACTER VINELANDII, OXIDIZED STATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Peters, J.W, Stowell, M.H.B, Soltis, S.M, Day, M.W, Kim, J, Rees, D.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-dependent structural changes in the nitrogenase P-cluster.
Biochemistry, 36, 1997
|
|
1CVX
 
 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1CXA
 
 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C, IMIDAZOLE | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-02-14 | | Release date: | 1995-07-10 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1CXC
 
 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-02-14 | | Release date: | 1995-09-15 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1CVY
 
 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYPYPYBETADP)2 BOUND TO CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1D8G
 
 | | ULTRAHIGH RESOLUTION CRYSTAL STRUCTURE OF B-DNA DECAMER D(CCAGTACTGG) | | Descriptor: | 5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*GP*)-3', CALCIUM ION | | Authors: | Kielkopf, C.L, Ding, S, Kuhn, P, Rees, D.C. | | Deposit date: | 1999-10-23 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Conformational flexibility of B-DNA at 0.74 A resolution: d(CCAGTACTGG)(2).
J.Mol.Biol., 296, 2000
|
|
1CP2
 
 | | NITROGENASE IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Schlessman, J.L, Woo, D, Joshua-Tor, L, Howard, J.B, Rees, D.C. | | Deposit date: | 1998-05-11 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
