2CPS
 
 | | SOLUTION NMR STRUCTURES OF THE MAJOR COAT PROTEIN OF FILAMENTOUS BACTERIOPHAGE M13 SOLUBILIZED IN SODIUM DODECYL SULPHATE MICELLES, 25 LOWEST ENERGY STRUCTURES | | Descriptor: | M13 MAJOR COAT PROTEIN | | Authors: | Papavoine, C.H.M, Christiaans, B.E.C, Folmer, R.H.A, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the M13 major coat protein in detergent micelles: a basis for a model of phage assembly involving specific residues.
J.Mol.Biol., 282, 1998
|
|
2CPB
 
 | | SOLUTION NMR STRUCTURES OF THE MAJOR COAT PROTEIN OF FILAMENTOUS BACTERIOPHAGE M13 SOLUBILIZED IN DODECYLPHOSPHOCHOLINE MICELLES, 25 LOWEST ENERGY STRUCTURES | | Descriptor: | M13 MAJOR COAT PROTEIN | | Authors: | Papavoine, C.H.M, Christiaans, B.E.C, Folmer, R.H.A, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the M13 major coat protein in detergent micelles: a basis for a model of phage assembly involving specific residues.
J.Mol.Biol., 282, 1998
|
|
2GVA
 
 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
2GVB
 
 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
1YHB
 
 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
1YHA
 
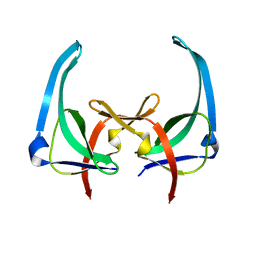 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
4RNF
 
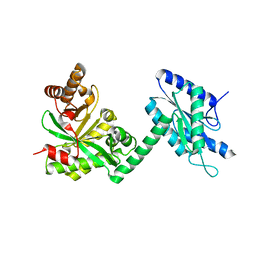 | |
4RNH
 
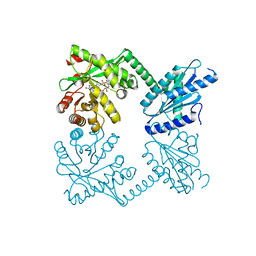 | | PaMorA tandem diguanylate cyclase - phosphodiesterase, c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
4RNJ
 
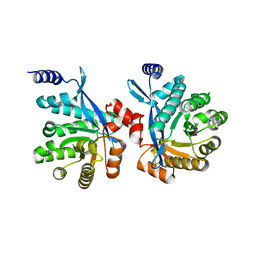 | | PaMorA phosphodiesterase domain, apo form | | Descriptor: | Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
4RNI
 
 | | PaMorA dimeric phosphodiesterase. apo form | | Descriptor: | Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
4V4L
 
 | | Structure of the Drosophila apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK, MAGNESIUM ION | | Authors: | Yuan, S, Topf, M, Akey, C.W, Ludtke, S.J. | | Deposit date: | 2010-10-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the Drosophila apoptosome at 6.9 angstrom resolution
Structure, 19, 2011
|
|
4V4N
 
 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
6TUT
 
 | | Cryo-EM structure of the RNA Polymerase III-Maf1 complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-02-19 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for RNA polymerase III transcription repression by Maf1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4V3E
 
 | | The CIDRa domain from IT4var07 PfEMP1 bound to endothelial protein C receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOTHELIAL PROTEIN C RECEPTOR, ... | | Authors: | Lau, C.K.Y, Turner, L, Jespersen, J.S, Lowe, E.D, Petersen, B, Wang, C.W, Petersen, J.E.V, Lusingu, J, Theander, T.G, Lavstsen, T, Higgins, M.K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Conservation Despite Huge Sequence Diversity Allows Epcr Binding by the Pfemp1 Family Implicated in Severe Childhood Malaria.
Cell Host Microbe., 17, 2015
|
|
4URS
 
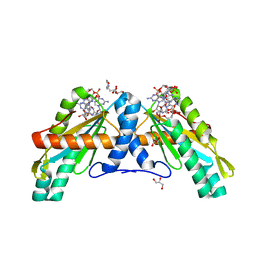 | | Crystal Structure of GGDEF domain from T.maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE, ... | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
4URQ
 
 | | Crystal Structure of GGDEF domain (I site mutant) from T.maritima | | Descriptor: | DIGUANYLATE CYCLASE | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swamianthan, K, Lescar, J. | | Deposit date: | 2014-07-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
4URG
 
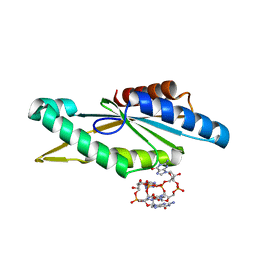 | | Crystal Structure of GGDEF domain from T.maritima (active-like dimer) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
4WLR
 
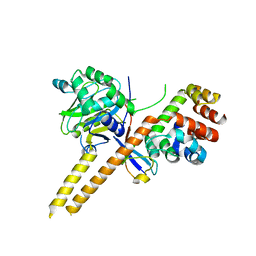 | | Crystal Structure of mUCH37-hRPN13 CTD-hUb complex | | Descriptor: | Polyubiquitin-B, Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WLP
 
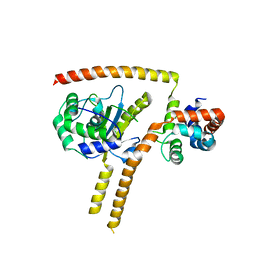 | | Crystal structure of UCH37-NFRKB Inhibited Deubiquitylating Complex | | Descriptor: | Nuclear factor related to kappa-B-binding protein, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
3PHP
 
 | | STRUCTURE OF THE 3' HAIRPIN OF THE TYMV PSEUDOKNOT: PREFORMATION IN RNA FOLDING | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*GP*AP*GP*GP*GP*UP*CP*AP*UP*CP*GP*GP*AP*AP*CP*CP*A) -3') | | Authors: | Kolk, M.H, Van Der Graaf, M, Wijmenga, S.S, Pleij, C.W.A, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the 3'-hairpin of the TYMV pseudoknot: preformation in RNA folding.
EMBO J., 17, 1998
|
|
1HIP
 
 | | TWO-ANGSTROM CRYSTAL STRUCTURE OF OXIDIZED CHROMATIUM HIGH POTENTIAL IRON PROTEIN | | Descriptor: | HIGH POTENTIAL IRON PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Carterjunior, C.W, Kraut, J, Freer, S.T, Xuong, N.-H, Alden, R.A, Bartsch, R.G. | | Deposit date: | 1975-04-01 | | Release date: | 1976-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein.
J.Biol.Chem., 249, 1974
|
|
3FI0
 
 | | Crystal Structure Analysis of B. stearothermophilus Tryptophanyl-tRNA Synthetase Complexed with Tryptophan, AMP, and Inorganic Phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, TRYPTOPHAN, ... | | Authors: | Laowanapiban, P, Kapustina, M, Vonrhein, C, Delarue, M, Koehl, P, Carter Jr, C.W. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3FHJ
 
 | | Independent saturation of three TrpRS subsites generates a partially-assembled state similar to those observed in molecular simulations | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, TRYPTOPHAN, ... | | Authors: | Laowanapiban, P, Kapustina, M, Vonrhein, C, Delarue, M, Koehl, P, Carter Jr, C.W. | | Deposit date: | 2008-12-09 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3H6Z
 
 | |
