1H6M
 
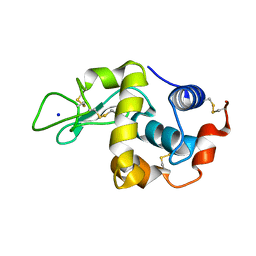 | | Covalent glycosyl-enzyme intermediate of hen egg white lysozyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose, LYSOZYME C, SODIUM ION | | Authors: | Vocadlo, D.J, Davies, G.J, Laine, R, Withers, S.G. | | Deposit date: | 2001-06-19 | | Release date: | 2001-08-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Catalysis by Hen Egg-White Lysozyme Proceeds Via a Covalent Intermediate
Nature, 412, 2001
|
|
8P0L
 
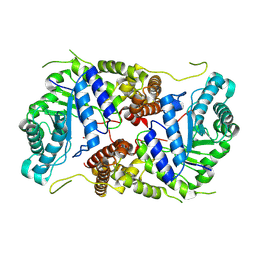 | | Crystal structure of human O-GlcNAcase in complex with an S-linked CKII peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Protein O-GlcNAcase, ... | | Authors: | Males, A, Davies, G.J, Calvelo, M, Alteen, M.G, Vocadlo, D.J, Rovira, C. | | Deposit date: | 2023-05-10 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human O -GlcNAcase Uses a Preactivated Boat-skew Substrate Conformation for Catalysis. Evidence from X-ray Crystallography and QM/MM Metadynamics.
Acs Catalysis, 13, 2023
|
|
1PX8
 
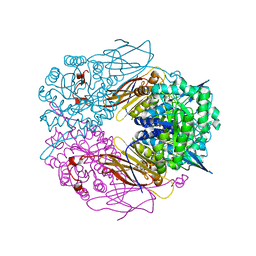 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
7LXG
 
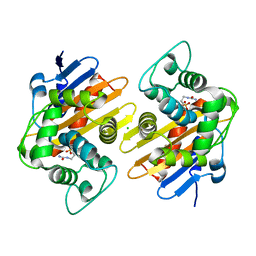 | |
1UHV
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
4AK5
 
 | | Native crystal structure of BpGH117 | | Descriptor: | 1,2-ETHANEDIOL, ANHYDRO-ALPHA-L-GALACTOSIDASE, CHLORIDE ION, ... | | Authors: | Hehemann, J.H, Smyth, L, Yadav, A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-14 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Keystone Enzyme in Agar Hydrolysis Provides Insight Into the Degradation (of a Polysaccharide from) Red Seaweeds.
J.Biol.Chem., 287, 2012
|
|
4AK6
 
 | | BpGH117_H302E mutant glycoside hydrolase | | Descriptor: | ANHYDRO-ALPHA-L-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Hehemann, J.H, Smyth, L, Yadav, A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-21 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of Keystone Enzyme in Agar Hydrolysis Provides Insight Into the Degradation (of a Polysaccharide from) Red Seaweeds.
J.Biol.Chem., 287, 2012
|
|
1ZUN
 
 | | Crystal Structure of a GTP-Regulated ATP Sulfurylase Heterodimer from Pseudomonas syringae | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Mougous, J.D, Lee, D.H, Hubbard, S.C, Schelle, M.W, Vocadlo, D.J, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for g protein control of the prokaryotic ATP sulfurylase.
Mol.Cell, 21, 2006
|
|
1JAK
 
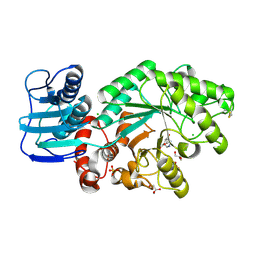 | | Streptomyces plicatus beta-N-acetylhexosaminidase in Complex with (2R,3R,4S,5R)-2-acetamido-3,4-dihydroxy-5-hydroxymethyl-piperidinium chloride (IFG) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Mark, B.L, Vocadlo, D.J, Zhao, D, Knapp, S, Withers, S.G, James, M.N. | | Deposit date: | 2001-05-30 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 beta-N-acetylhexosaminidase inhibitor.
J.Biol.Chem., 276, 2001
|
|
2CHN
 
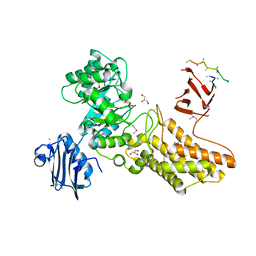 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity- NAG-thiazoline complex | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
2CHO
 
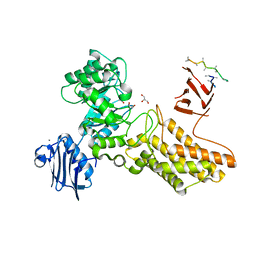 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity | | Descriptor: | ACETATE ION, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
5M7R
 
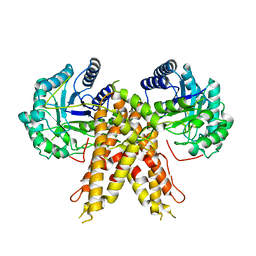 | | Structure of human O-GlcNAc hydrolase | | Descriptor: | Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
5M7T
 
 | | Structure of human O-GlcNAc hydrolase with PugNAc type inhibitor | | Descriptor: | (5R,6R,7R,8S)-8-(ACETYLAMINO)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-N-PHENYL-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDINE-2-CARBOXAMIDE, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
5M7S
 
 | | Structure of human O-GlcNAc hydrolase with bound transition state analog ThiametG | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
5M7U
 
 | | Structure of human O-GlcNAc hydrolase with new iminocyclitol type inhibitor | | Descriptor: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-[3-[3-(trifluoromethyl)phenyl]propyl]pyrrolidin-2-yl]-~{N}-methyl-ethanamide, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
7OU6
 
 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | Protein O-GlcNAcase, ~{N}-[(3~{Z},6~{S},7~{R},8~{R},8~{a}~{S})-7,8-bis(oxidanyl)-3-(phenylmethyl)imino-1,5,6,7,8,8~{a}-hexahydro-[1,3]thiazolo[3,4-a]pyridin-6-yl]ethanamide | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
7OU8
 
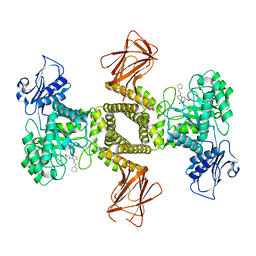 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, O-GlcNAcase BT_4395, ... | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
1M04
 
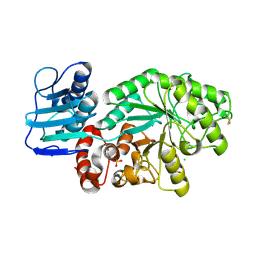 | | Mutant Streptomyces plicatus beta-hexosaminidase (D313N) in complex with product (GlcNAc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Williams, S.J, Mark, B.L, Vocadlo, D.J, James, M.N.G, Withers, S.G. | | Deposit date: | 2002-06-11 | | Release date: | 2002-12-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical
role in substrate-assisted catalysis by orienting the 2-acetamido group
and stabilizing the transition state.
J.Biol.Chem., 277, 2002
|
|
1M03
 
 | | Mutant Streptomyces plicatus beta-hexosaminidase (D313A) in complex with product (GlcNAc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Williams, S.J, Mark, B.L, Vocadlo, D.J, James, M.N.G, Withers, S.G. | | Deposit date: | 2002-06-11 | | Release date: | 2002-12-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical
role in substrate-assisted catalysis by orienting the 2-acetamido group
and stabilizing the transition state.
J.Biol.Chem., 277, 2002
|
|
1M01
 
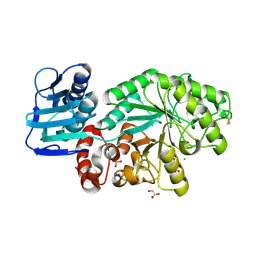 | | Wildtype Streptomyces plicatus beta-hexosaminidase in complex with product (GlcNAc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | J Williams, S, Mark, B.L, Vocadlo, D.J, James, M.N.G, Withers, S.G. | | Deposit date: | 2002-06-11 | | Release date: | 2003-01-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical
role in substrate-assisted catalysis by orienting the 2-acetamido group
and stabilizing the transition state.
J.Biol.Chem., 277, 2002
|
|
5FL0
 
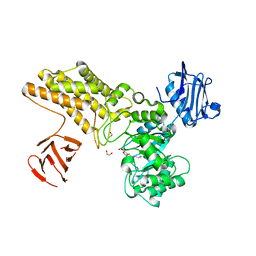 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-(butylamino)-5-(hydroxymethyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FL1
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FQF
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQH
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FR0
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
