4RCR
 
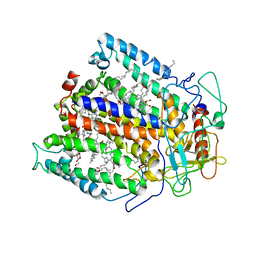 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
2Z9H
 
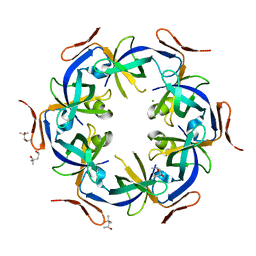 | | Ethanolamine utilization protein, EutN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, Ethanolamine utilization protein eutN | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The crystal structure of ethanolamine utilization protein EutN from E. coli
To be Published
|
|
1YAC
 
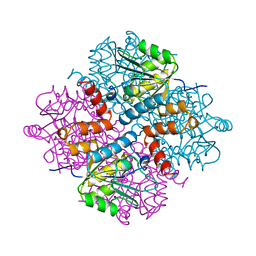 | |
6NHT
 
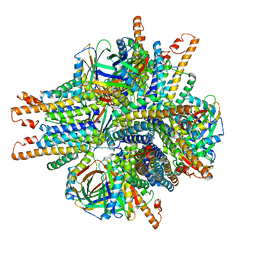 | |
6NHV
 
 | |
1CYJ
 
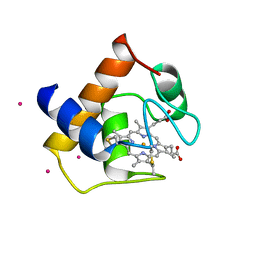 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1CYI
 
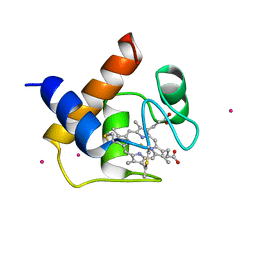 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1DOF
 
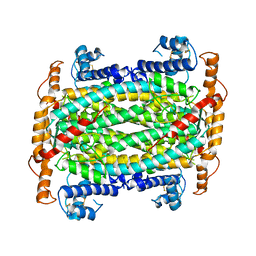 | | THE CRYSTAL STRUCTURE OF ADENYLOSUCCINATE LYASE FROM PYROBACULUM AEROPHILUM: INSIGHTS INTO THERMAL STABILITY AND HUMAN PATHOLOGY | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T.O, Goedken, E, Dixon, J.E, Marqusee, S. | | Deposit date: | 1999-12-20 | | Release date: | 2001-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
6XPH
 
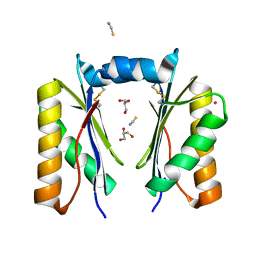 | | CutR dimer with domain swap | | Descriptor: | Ethanolamine utilization protein EutS, GLYCEROL, POTASSIUM ION, ... | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O, Nie, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6XPK
 
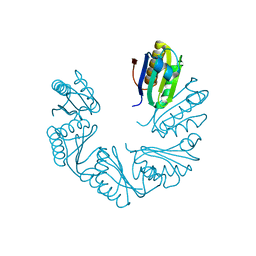 | | CutR Screw, form 1 with 41.9 angstrom pitch | | Descriptor: | Ethanolamine utilization protein EutS | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6XPL
 
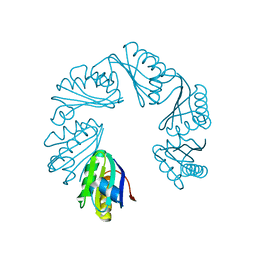 | | CutR Screw, form 2 with 33.8 angstrom pitch | | Descriptor: | Ethanolamine utilization protein EutS | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6XPI
 
 | | CutR flat hexamer, form 1 | | Descriptor: | Ethanolamine utilization protein EutS | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6XPJ
 
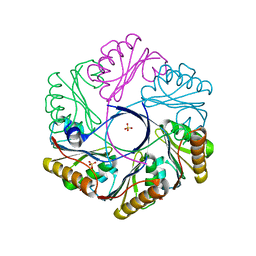 | | CutR flat hexamer, form 2 | | Descriptor: | Ethanolamine utilization protein EutS, SULFATE ION | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
1F1F
 
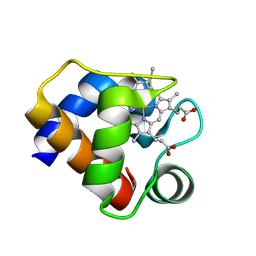 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1F1C
 
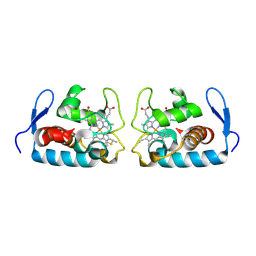 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | Descriptor: | CYTOCHROME C549, HEME C | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
5UI2
 
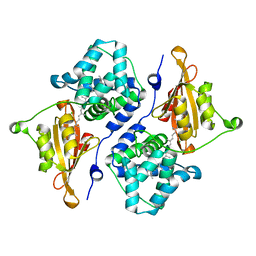 | | CRYSTAL STRUCTURE OF ORANGE CAROTENOID PROTEIN | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, CHLORIDE ION, Orange carotenoid-binding protein, ... | | Authors: | KERFELD, C.A, SAWAYA, M.R, VISHNU, B, KROGMANN, D, YEATES, T.O. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a cyanobacterial water-soluble carotenoid binding protein.
Structure, 11, 2003
|
|
8UJA
 
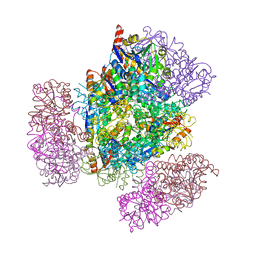 | |
7MPW
 
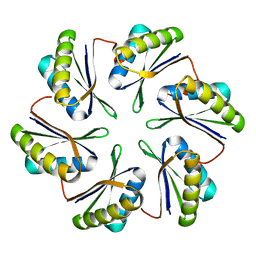 | | CmcB from Type II Cut MCP | | Descriptor: | BMC domain-containing protein | | Authors: | Ochoa, J.M, Marshall, J.D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MGP
 
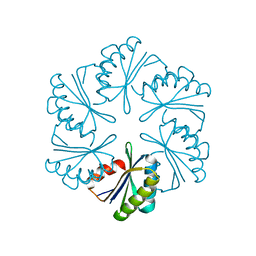 | | CmcA from Type II Cut MCP | | Descriptor: | BMC domain-containing protein | | Authors: | Ochoa, J.M, Escoto, X, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-04-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MN4
 
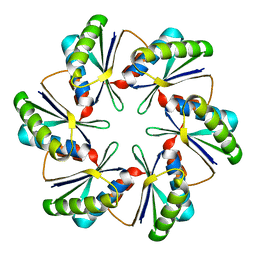 | | CmcB E35G mutant from Type II Cut MCP | | Descriptor: | BMC domain-containing protein | | Authors: | Ochoa, J.M, Mijares, O, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MPV
 
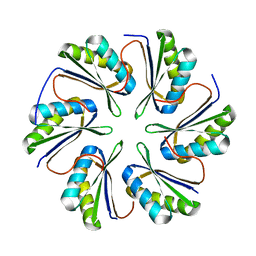 | | CmcC from Type II Cut MCP | | Descriptor: | BMC domain-containing protein | | Authors: | Ochoa, J.M, Mijares, O, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MPX
 
 | | CmcC E36G mutant from Type II Cut MCP | | Descriptor: | BMC domain-containing protein, PHOSPHATE ION | | Authors: | Ochoa, J.M, Acosta, A.A, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MMX
 
 | | CutN from Type I Cut MCP | | Descriptor: | Propanediol utilization protein PduA, SULFATE ION | | Authors: | Ochoa, J.M, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8G47
 
 | | KRAS G12C complex with GDP and AMG 510 imaged on a cryo-EM imaging scaffold | | Descriptor: | AMG 510 (bound form), GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G3K
 
 | |
