1DDJ
 
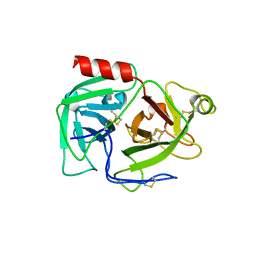 | | CRYSTAL STRUCTURE OF HUMAN PLASMINOGEN CATALYTIC DOMAIN | | Descriptor: | PLASMINOGEN | | Authors: | Wang, X, Terzyan, S, Tang, J, Loy, J, Lin, X, Zhang, X. | | Deposit date: | 1999-11-10 | | Release date: | 2000-02-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human plasminogen catalytic domain undergoes an unusual conformational change upon activation.
J.Mol.Biol., 295, 2000
|
|
1AKN
 
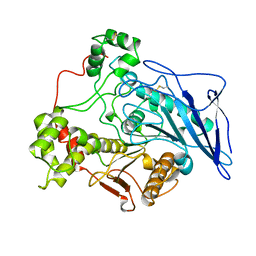 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-05-23 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
1AQL
 
 | |
1BML
 
 | |
1C4P
 
 | | BETA DOMAIN OF STREPTOKINASE | | Descriptor: | PROTEIN (STREPTOKINASE) | | Authors: | Wang, X, Zhang, X.C. | | Deposit date: | 1999-09-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of streptokinase beta-domain.
FEBS Lett., 459, 1999
|
|
1G2E
 
 | |
1FXL
 
 | |
7A1V
 
 | |
1IB2
 
 | |
3TKR
 
 | | Crystal structure of full-length human peroxiredoxin 4 with T118E mutation | | Descriptor: | Peroxiredoxin-4 | | Authors: | Wang, X, Wang, L, Wang, X, Sun, F, Wang, C.-C. | | Deposit date: | 2011-08-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4
Biochem.J., 2011
|
|
3TKQ
 
 | | Crystal structure of full-length human peroxiredoxin 4 with mixed conformation | | Descriptor: | Peroxiredoxin-4 | | Authors: | Wang, X, Wang, L, Wang, X, Sun, F, Wang, C.-C. | | Deposit date: | 2011-08-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4
Biochem.J., 2011
|
|
3TKS
 
 | | Crystal structure of full-length human peroxiredoxin 4 in three different redox states | | Descriptor: | PEROXIDE ION, Peroxiredoxin-4 | | Authors: | Wang, X, Wang, L, Wang, X, Sun, F, Wang, C.-C. | | Deposit date: | 2011-08-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4
Biochem.J., 2011
|
|
3TKP
 
 | | Crystal structure of full-length human peroxiredoxin 4 in the reduced form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Wang, X, Wang, L, Wang, X, Sun, F, Wang, C.-C. | | Deposit date: | 2011-08-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4
Biochem.J., 2011
|
|
3VBS
 
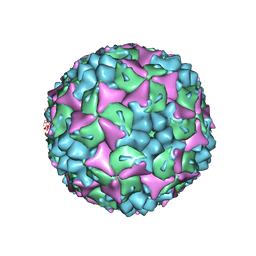 | | Crystal structure of human Enterovirus 71 | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBU
 
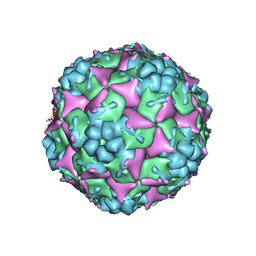 | | Crystal structure of empty human Enterovirus 71 particle | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBH
 
 | | Crystal structure of formaldehyde treated human enterovirus 71 (space group R32) | | Descriptor: | CHLORIDE ION, Genome Polyprotein, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBR
 
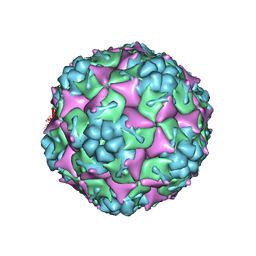 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (room temperature) | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBF
 
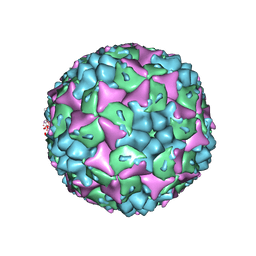 | | Crystal structure of formaldehyde treated human Enterovirus 71 (space group I23) | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Genome Polyprotein, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBO
 
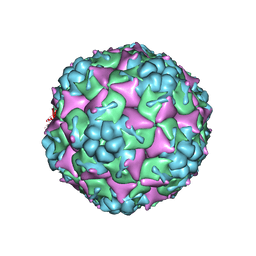 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (cryo at 100K) | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3NFC
 
 | |
7F16
 
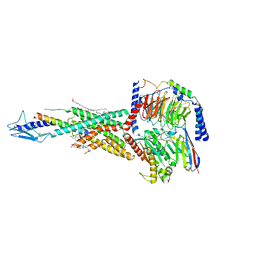 | | Cryo-EM structure of parathyroid hormone receptor type 2 in complex with a tuberoinfundibular peptide of 39 residues and G protein | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X, Cheng, X, Zhao, L.H, Wang, Y, Ye, C, Zou, X, Dai, A, Cong, Z, Chen, J, Zhou, Q, Xia, T, Jiang, H, Xu, H.E, Yang, D, Wang, M.W. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1M8W
 
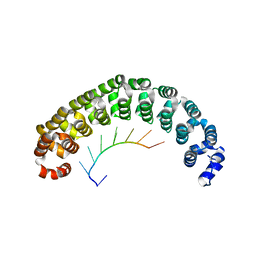 | | CRYSTAL STRUCTURE OF THE PUMILIO-HOMOLOGY DOMAIN FROM HUMAN PUMILIO1 IN COMPLEX WITH NRE1-19 RNA | | Descriptor: | 5'-R(P*UP*GP*UP*AP*UP*AP*U)-3', 5'-R(P*UP*GP*UP*CP*CP*AP*G)-3', 5'-R(P*UP*UP*GP*UP*AP*UP*AP*U)-3', ... | | Authors: | Wang, X, McLachlan, J, Zamore, P.D, Hall, T.M.T. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MODULAR RECOGNITION OF RNA BY A HUMAN PUMILIO-HOMOLOGY DOMAIN
CELL(CAMBRIDGE,MASS.), 110, 2002
|
|
1M8Y
 
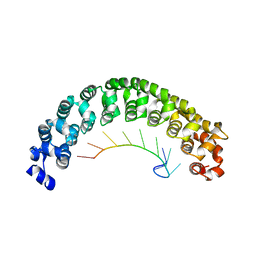 | | CRYSTAL STRUCTURE OF THE PUMILIO-HOMOLOGY DOMAIN FROM HUMAN PUMILIO1 IN COMPLEX WITH NRE2-10 RNA | | Descriptor: | 5'-R(P*AP*UP*UP*GP*UP*AP*CP*AP*UP*A)-3', Pumilio 1 | | Authors: | Wang, X, McLachlan, J, Zamore, P.D, Hall, T.M.T. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MODULAR RECOGNITION OF RNA BY A HUMAN PUMILIO-HOMOLOGY DOMAIN
CELL(CAMBRIDGE,MASS.), 110, 2002
|
|
1M8X
 
 | | CRYSTAL STRUCTURE OF THE PUMILIO-HOMOLOGY DOMAIN FROM HUMAN PUMILIO1 IN COMPLEX WITH NRE1-14 RNA | | Descriptor: | 5'-R(P*UP*GP*UP*AP*UP*AP*U)-3', 5'-R(P*UP*UP*GP*UP*AP*UP*AP*U)-3', Pumilio 1 | | Authors: | Wang, X, McLachlan, J, Zamore, P.D, Hall, T.M.T. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MODULAR RECOGNITION OF RNA BY A HUMAN PUMILIO-HOMOLOGY DOMAIN
CELL(CAMBRIDGE,MASS.), 110, 2002
|
|
1M8Z
 
 | |
