5AT1
 
 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
4AT1
 
 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
6AT1
 
 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
2AT2
 
 | |
1ACM
 
 | | ARGININE 54 IN THE ACTIVE SITE OF ESCHERICHIA COLI ASPARTATE TRANSCARBAMOYLASE IS CRITICAL FOR CATALYSIS: A SITE-SPECIFIC MUTAGENESIS, NMR AND X-RAY CRYSTALLOGRAPHY STUDY | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ASPARTATE CARBAMOYLTRANSFERASE, CATALYTIC CHAIN, ... | | Authors: | Stevens, R.C, Kantrowitz, E.R, Lipscomb, W.N. | | Deposit date: | 1992-07-08 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Arginine 54 in the active site of Escherichia coli aspartate transcarbamoylase is critical for catalysis: a site-specific mutagenesis, NMR, and X-ray crystallographic study.
Protein Sci., 1, 1992
|
|
3BTA
 
 | |
3CZO
 
 | |
2NZ9
 
 | |
2NYY
 
 | |
2PAH
 
 | |
1FL3
 
 | |
1HTZ
 
 | | CRYSTAL STRUCTURE OF TEM52 BETA-LACTAMASE | | Descriptor: | BETA-LACTAMASE MUTANT TEM52 | | Authors: | Stevens, R.C, Orencia, M.C. | | Deposit date: | 2001-01-03 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predicting the emergence of antibiotic resistance by directed evolution and structural analysis.
Nat.Struct.Biol., 8, 2001
|
|
1PAH
 
 | |
7AT1
 
 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ... | | Authors: | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | Deposit date: | 1989-09-22 | | Release date: | 1990-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
1AJ7
 
 | | IMMUNOGLOBULIN 48G7 GERMLINE FAB ANTIBODY COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID. AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY | | Descriptor: | 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID, IMMUNOGLOBULIN 48G7 FAB (HEAVY CHAIN), IMMUNOGLOBULIN 48G7 FAB (LIGHT CHAIN) | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1997-05-15 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the evolution of an antibody combining site.
Science, 276, 1997
|
|
5PAH
 
 | |
8AT1
 
 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | Deposit date: | 1989-09-22 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
7DTY
 
 | | Structural basis of ligand selectivity conferred by the human glucose-dependent insulinotropic polypeptide receptor | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhang, C, Zhou, Q.T, Hang, K.N, Zou, X.Y, Chen, Y, Wu, F, Rao, Q.D, Dai, A.T, Yin, W.C, Shen, D.D, Zhang, Y, Xia, T, Stevens, R.C, Xu, H.E, Yang, D.H, Zhao, L.H, Wang, M.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-04 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor.
Elife, 10, 2021
|
|
6W25
 
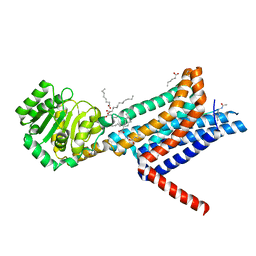 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
1C7J
 
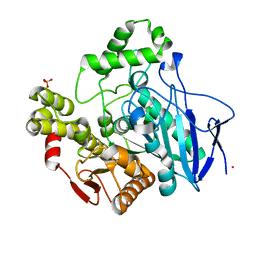 | | PNB ESTERASE 56C8 | | Descriptor: | POTASSIUM ION, PROTEIN (PARA-NITROBENZYL ESTERASE), SULFATE ION | | Authors: | Spiller, B, Gershenson, A, Arnold, F, Stevens, R.C. | | Deposit date: | 2000-02-21 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1A4J
 
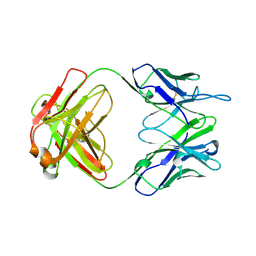 | | DIELS ALDER CATALYTIC ANTIBODY GERMLINE PRECURSOR | | Descriptor: | IMMUNOGLOBULIN, DIELS ALDER CATALYTIC ANTIBODY (HEAVY CHAIN), DIELS ALDER CATALYTIC ANTIBODY (LIGHT CHAIN) | | Authors: | Spiller, B.W, Romesburg, F.E, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Immunological origins of binding and catalysis in a Diels-Alderase antibody.
Science, 279, 1998
|
|
1A4K
 
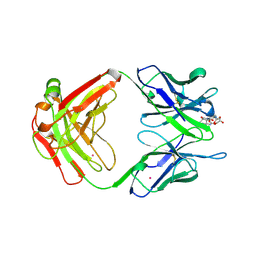 | | DIELS ALDER CATALYTIC ANTIBODY WITH TRANSITION STATE ANALOGUE | | Descriptor: | ANTIBODY FAB, CADMIUM ION, [4-(4-ACETYLAMINO-PHENYL)-3,5-DIOXO-4-AZA-TRICYCLO[5.2.2.0 2,6]UNDEC-1-YLCARBAMOYLOXY]-ACETIC ACID | | Authors: | Spiller, B.W, Romesburg, F.E, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Immunological origins of binding and catalysis in a Diels-Alderase antibody.
Science, 279, 1998
|
|
1AXS
 
 | | MATURE OXY-COPE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | (1S,2S,5S)2-(4-GLUTARIDYLBENZYL)-5-PHENYL-1-CYCLOHEXANOL, CADMIUM ION, OXY-COPE CATALYTIC ANTIBODY | | Authors: | Mundorff, E.C, Ulrich, H.D, Stevens, R.C. | | Deposit date: | 1997-10-20 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interplay between binding energy and catalysis in the evolution of a catalytic antibody.
Nature, 389, 1997
|
|
6PAH
 
 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND L-DOPA (3,4-DIHYDROXYPHENYLALANINE) INHIBITOR | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, FE (III) ION, PHENYLALANINE 4-MONOOXYGENASE | | Authors: | Erlandsen, H, Flatmark, T, Stevens, R.C. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
6PT2
 
 | | Crystal structure of the active delta opioid receptor in complex with the peptide agonist KGCHM07 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Delta opioid receptor, ... | | Authors: | Claff, T, Yu, J, Blais, V, Patel, N, Martin, C, Wu, L, Han, G.W, Holleran, B.J, Van der Poorten, O, Hanson, M.A, Sarret, P, Gendron, L, Cherezov, V, Katritch, V, Ballet, S, Liu, Z, Muller, C.E, Stevens, R.C. | | Deposit date: | 2019-07-14 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Sci Adv, 5, 2019
|
|
