1GCO
 
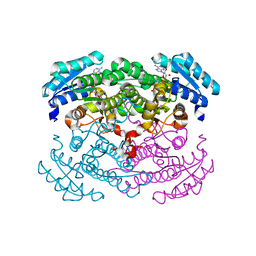 | | CRYSTAL STRUCTURE OF GLUCOSE DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | GLUCOSE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glucose dehydrogenase from Bacillus megaterium IWG3 at 1.7 A resolution.
J.Biochem., 129, 2001
|
|
1GEE
 
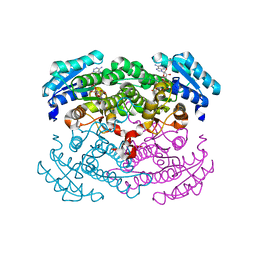 | | Crystal structure of glucose dehydrogenase mutant Q252L complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-07 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
1G6K
 
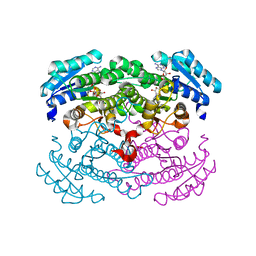 | | Crystal structure of glucose dehydrogenase mutant E96A complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
5H5L
 
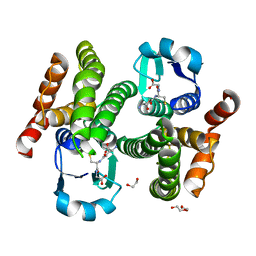 | | Structure of prostaglandin synthase D of Nilaparvata lugens | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, S, Nakagawa, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular structure of a prostaglandin D synthase requiring glutathione from the brown planthopper, Nilaparvata lugens
Biochem. Biophys. Res. Commun., 492, 2017
|
|
5X7Y
 
 | | Crystal Structure of the Dog Lipocalin Allergen Can f 6 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipocalin-Can f 6 allergen | | Authors: | Yamamoto, K, Otani, T, Sugiura, K, Nakatsuji, M, Nishimura, S, Inui, T. | | Deposit date: | 2017-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the dog allergen Can f 6 and structure-based implications of its cross-reactivity with the cat allergen Fel d 4.
Sci Rep, 9, 2019
|
|
5ZFG
 
 | | Crystal structure of a diazinon-metabolizing glutathione S-transferase in the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterisation of a diazinon-metabolising glutathione S-transferase in the silkworm Bombyx mori by X-ray crystallography and genome editing analysis.
Sci Rep, 8, 2018
|
|
7XHX
 
 | | Crystal structure of metallo-beta-lactamase IMP-6 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
7XHW
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
5AZ1
 
 | | Crystal structure of aldo-keto reductase (AKR2E5) complexed with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5AZ0
 
 | | Crystal structure of aldo-keto reductase (AKR2E5) of the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
3VPQ
 
 | | Crystal structure of Bombyx mori sigma-class glutathione transferase in complex with glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, Glutathione S-transferase sigma, ... | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A, Suzuki, M. | | Deposit date: | 2012-03-08 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure of a Bombyx mori sigma-class glutathione transferase exhibiting prostaglandin E synthase activity
Biochim.Biophys.Acta, 1830, 2013
|
|
3VPT
 
 | | Crystal structure of Bombyx mori sigma-class glutathione transferase in apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutathione S-transferase sigma, S-1,2-PROPANEDIOL, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Bombyx mori sigma-class glutathione transferase exhibiting prostaglandin E synthase activity
Biochim.Biophys.Acta, 1830, 2013
|
|
3A4A
 
 | | Crystal structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
3A47
 
 | |
3AJ7
 
 | | Crystal Structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
3AXH
 
 | | Crystal structure of isomaltase in complex with isomaltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
3AXI
 
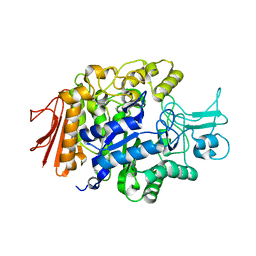 | | Crystal structure of isomaltase in complex with maltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
3VUR
 
 | |
3WCZ
 
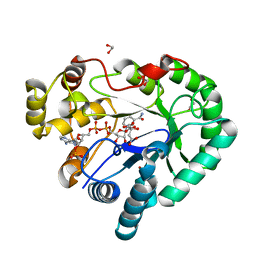 | | Crystal structure of Bombyx mori aldo-keto reductase (AKR2E4) in complex with NADP | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase 2E, CITRIC ACID, ... | | Authors: | Yamamoto, K, Wilson, D.K. | | Deposit date: | 2013-06-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification, characterization, and crystal structure of an aldo-keto reductase (AKR2E4) from the silkworm Bombyx mori.
Arch.Biochem.Biophys., 538, 2013
|
|
3WD6
 
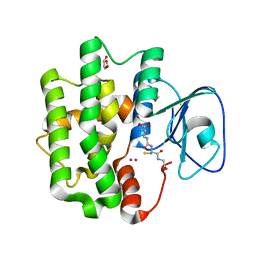 | | Crystal structure of Bombyx mori omega-class glutathione transferase in complex with GSH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Yamamoto, K, Suzuki, M, Higashiura, A, Nakagawa, A. | | Deposit date: | 2013-06-07 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of a Bombyx mori Omega-class glutathione transferase.
Biochem.Biophys.Res.Commun., 438, 2013
|
|
3WYW
 
 | | Structural characterization of catalytic site of a Nilaparvata lugens delta-class glutathione transferase | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A. | | Deposit date: | 2014-09-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of the catalytic site of a Nilaparvata lugens delta-class glutathione transferase.
Arch.Biochem.Biophys., 566C, 2014
|
|
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | Descriptor: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
6JXK
 
 | | Rb+-bound E2-MgF state of the gastric proton pump (Wild-type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Abe, K, Irie, K, Yamamoto, K. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | A single K + -binding site in the crystal structure of the gastric proton pump.
Elife, 8, 2019
|
|
6K0T
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-17 | | Descriptor: | 3-[(1~{E})-1-[8-[(8-chloranyl-2-cyclopropyl-imidazo[1,2-a]pyridin-3-yl)methyl]-3-fluoranyl-6~{H}-benzo[c][1]benzoxepin-11-ylidene]ethyl]-4~{H}-1,2,4-oxadiazol-5-one, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Suzuki, M, Yamamoto, K, Takahashi, Y, Saito, J. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-30 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Development of a novel class of peroxisome proliferator-activated receptor (PPAR) gamma ligands as an anticancer agent with a unique binding mode based on a non-thiazolidinedione scaffold.
Bioorg.Med.Chem., 27, 2019
|
|
3VK9
 
 | | Crystal structure of delta-class glutathione transferase from silkmoth | | Descriptor: | GLYCEROL, Glutathione S-transferase delta | | Authors: | Kakuta, Y, Usuda, K, Higashiura, A, Suzuki, M, Nakagawa, A, Kimura, M, Yamamoto, K. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for catalytic activity of a silkworm Delta-class glutathione transferase
Biochim.Biophys.Acta, 1820, 2012
|
|
