4TNC
 
 | |
1BPQ
 
 | |
1UNE
 
 | |
1TOP
 
 | |
1NCZ
 
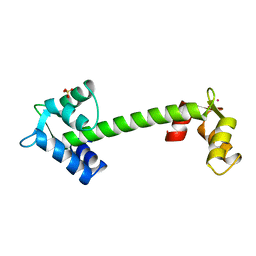 | | TROPONIN C | | Descriptor: | SULFATE ION, TERBIUM(III) ION, TROPONIN C | | Authors: | Sundaralingam, M, Rao, S.T. | | Deposit date: | 1996-06-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Mn, Cd and Tb metal complexes of troponin C.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1NCY
 
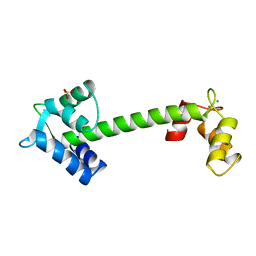 | | TROPONIN-C, COMPLEX WITH MANGANESE | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TROPONIN C | | Authors: | Sundaralingam, M, Rao, S.T. | | Deposit date: | 1996-06-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of Mn, Cd and Tb metal complexes of troponin C.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1NCX
 
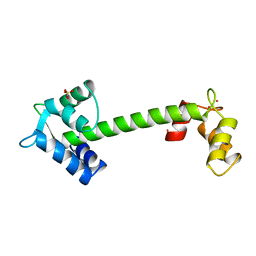 | | TROPONIN C | | Descriptor: | CADMIUM ION, SULFATE ION, TROPONIN C | | Authors: | Sundaralingam, M, Rao, S.T. | | Deposit date: | 1996-06-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Mn, Cd and Tb metal complexes of troponin C.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1FDK
 
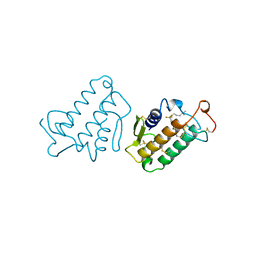 | | CARBOXYLIC ESTER HYDROLASE (PLA2-MJ33 INHIBITOR COMPLEX) | | Descriptor: | 1-DECYL-3-TRIFLUORO ETHYL-SN-GLYCERO-2-PHOSPHOMETHANOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sundaralingam, M. | | Deposit date: | 1997-09-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the complex of bovine pancreatic phospholipase A2 with the inhibitor 1-hexadecyl-3-(trifluoroethyl)-sn-glycero-2-phosphomethanol,.
Biochemistry, 36, 1997
|
|
1KVW
 
 | |
1KVY
 
 | |
1KVX
 
 | |
1MKV
 
 | |
1OSA
 
 | |
1MKT
 
 | |
1MKU
 
 | |
1MKS
 
 | |
1IRB
 
 | | CARBOXYLIC ESTER HYDROLASE | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sundaralingam, M. | | Deposit date: | 1997-08-13 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phospholipase A2 engineering. Deletion of the C-terminus segment changes substrate specificity and uncouples calcium and substrate binding at the zwitterionic interface.
Biochemistry, 35, 1996
|
|
1CLM
 
 | |
2BPP
 
 | |
5LYM
 
 | |
1TRA
 
 | | RESTRAINED REFINEMENT OF THE MONOCLINIC FORM OF YEAST PHENYLALANINE TRANSFER RNA. TEMPERATURE FACTORS AND DYNAMICS, COORDINATED WATERS, AND BASE-PAIR PROPELLER TWIST ANGLES | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Westhof, E, Sundaralingam, M. | | Deposit date: | 1986-05-16 | | Release date: | 1986-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of the monoclinic form of yeast phenylalanine transfer RNA. Temperature factors and dynamics, coordinated waters, and base-pair propeller twist angles.
Biochemistry, 25, 1986
|
|
1VTB
 
 | |
138D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-HEXAGONAL CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
137D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
116D
 
 | |
