5D2N
 
 | | Crystal structure of C25-NLV-HLA-A2 complex | | Descriptor: | ASN-LEU-VAL-PRO-MET-VAL-ALA-THR-VAL, Beta-2-microglobulin, C25 alpha, ... | | Authors: | Mariuzza, R.A, Yang, X. | | Deposit date: | 2015-08-05 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural Basis for Clonal Diversity of the Public T Cell Response to a Dominant Human Cytomegalovirus Epitope.
J.Biol.Chem., 290, 2015
|
|
1NBY
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K96A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NDG
 
 | | Crystal structure of Fab fragment of antibody HyHEL-8 complexed with its antigen lysozyme | | Descriptor: | ACETIC ACID, Lysozyme C, antibody kappa light chain, ... | | Authors: | Mariuzza, R.A, Li, Y, Li, H, Yang, F, Smith-Gill, S.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-06-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of the maturation of an antibody response to a protein antigen
Nat.Struct.Biol., 10, 2003
|
|
1NBZ
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K97A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NDM
 
 | |
1DDH
 
 | |
8SR0
 
 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 local refined | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
8SO3
 
 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
7R6J
 
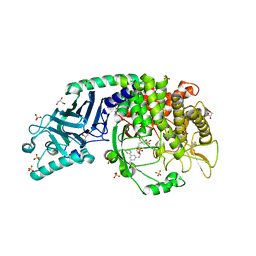 | | Co-crystal structure of Chaetomium glucosidase with compound 1 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[3-methyl-5-(pyrimidin-2-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
3T0E
 
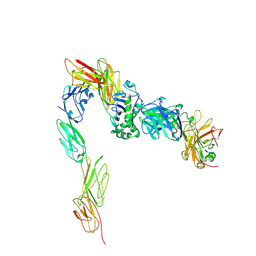 | | Crystal structure of a complete ternary complex of T cell receptor, peptide-MHC and CD4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-4 beta chain, ... | | Authors: | Yin, Y, Mariuzza, R.A. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1AC6
 
 | |
1A2Y
 
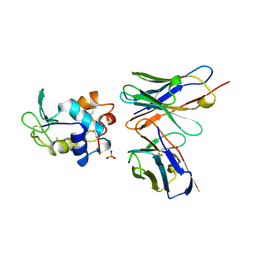 | | HEN EGG WHITE LYSOZYME, D18A MUTANT, IN COMPLEX WITH MOUSE MONOCLONAL ANTIBODY D1.3 | | Descriptor: | IGG1-KAPPA D1.3 FV (HEAVY CHAIN), IGG1-KAPPA D1.3 FV (LIGHT CHAIN), LYSOZYME, ... | | Authors: | Tsuchiya, D, Mariuzza, R.A. | | Deposit date: | 1998-01-13 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A mutational analysis of binding interactions in an antigen-antibody protein-protein complex.
Biochemistry, 37, 1998
|
|
5TEZ
 
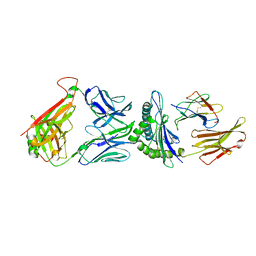 | | TCR F50 recgonizing M1-HLA-A2 | | Descriptor: | Beta-2-microglobulin, GLY-ILE-LEU-GLY-PHE-VAL-PHE-THR-LEU, HLA class I histocompatibility antigen, ... | | Authors: | Yang, X, Mariuzza, R.A. | | Deposit date: | 2016-09-23 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for clonal diversity of the human T-cell response to a dominant influenza virus epitope.
J. Biol. Chem., 292, 2017
|
|
4E41
 
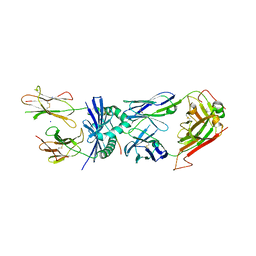 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
4E42
 
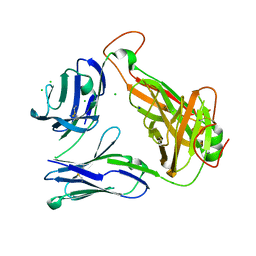 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | CHLORIDE ION, NITRATE ION, SODIUM ION, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
1BEC
 
 | |
4K79
 
 | | Recognition of the Thomsen-Friedenreich Antigen by a Lamprey Variable Lymphocyte Receptor | | Descriptor: | Variable lymphocyte receptor, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Luo, M, Velikovsky, C.A, Yang, X.B, Mariuzza, R.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the thomsen-friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
J.Biol.Chem., 288, 2013
|
|
4K5U
 
 | | Recognition of the BG-H Antigen by a Lamprey Variable Lymphocyte Receptor | | Descriptor: | Variable lymphocyte receptor, beta-D-galactopyranose | | Authors: | Luo, M, Velikovsky, C.A, Yang, X.B, Mariuzza, R.A. | | Deposit date: | 2013-04-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Recognition of the thomsen-friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
J.Biol.Chem., 288, 2013
|
|
2PTT
 
 | |
2PTU
 
 | | Structure of NK cell receptor 2B4 (CD244) | | Descriptor: | Natural killer cell receptor 2B4 | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
2PTV
 
 | | Structure of NK cell receptor ligand CD48 | | Descriptor: | CD48 antigen | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
3L6F
 
 | |
3M19
 
 | |
3MC0
 
 | | Crystal Structure of Staphylococcal Enterotoxin G (SEG) in Complex with a Mouse T-cell Receptor beta Chain | | Descriptor: | ACETATE ION, Enterotoxin SEG, variable beta 8.2 mouse T cell receptor | | Authors: | Fernandez, M.M, Cho, S, Robinson, H, Mariuzza, R.A, Malchiodi, E.L. | | Deposit date: | 2010-03-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcal enterotoxin G (SEG) in complex with a mouse T-cell receptor {beta} chain.
J.Biol.Chem., 286, 2011
|
|
3M18
 
 | |
