1BLP
 
 | |
5TNC
 
 | |
3BLM
 
 | |
1KBL
 
 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | AMMONIUM ION, PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C, Liu, S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis
Biochemistry, 41, 2002
|
|
1DIK
 
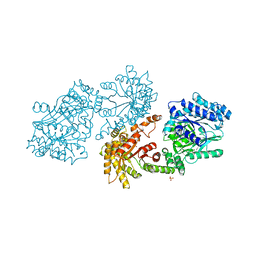 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C.H. | | Deposit date: | 1995-12-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2HPR
 
 | |
7LYZ
 
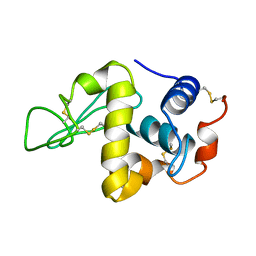 | | PROTEIN MODEL BUILDING BY THE USE OF A CONSTRAINED-RESTRAINED LEAST-SQUARES PROCEDURE | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Moult, J, Yonath, A, Sussman, J, Herzberg, O, Podjarny, A, Traub, W. | | Deposit date: | 1977-05-06 | | Release date: | 1977-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Model Building by the Use of a Constrained-Restrained Least-Squares Procedure
J.Appl.Crystallogr., 16, 1983
|
|
4QT8
 
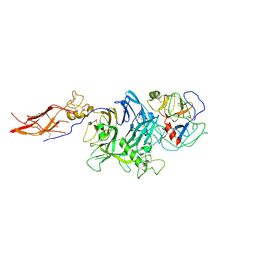 | | Crystal Structure of RON Sema-PSI-IPT1 extracellular domains in complex with MSP beta-chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor-like protein, Macrophage-stimulating protein receptor, ... | | Authors: | Herzberg, O, Chao, K.L. | | Deposit date: | 2014-07-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the binding specificity of human Recepteur d'Origine Nantais (RON) receptor tyrosine kinase to macrophage-stimulating protein.
J.Biol.Chem., 289, 2014
|
|
1GPR
 
 | |
1KC7
 
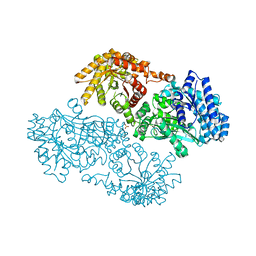 | | Pyruvate Phosphate Dikinase with Bound Mg-phosphonopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHONOPYRUVATE, SULFATE ION, ... | | Authors: | Chen, C.C, Howard, A, Herzberg, O. | | Deposit date: | 2001-11-07 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis
Biochemistry, 41, 2002
|
|
3LYE
 
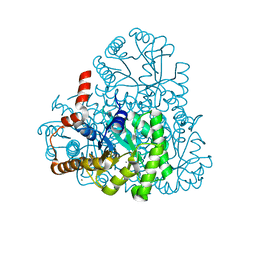 | | Crystal structure of oxaloacetate acetylhydrolase | | Descriptor: | CALCIUM ION, Oxaloacetate acetyl hydrolase | | Authors: | Herzberg, O, Chen, C. | | Deposit date: | 2010-02-26 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
J.Biol.Chem., 285, 2010
|
|
3M0K
 
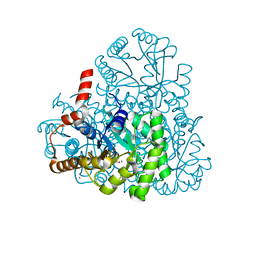 | |
3M0J
 
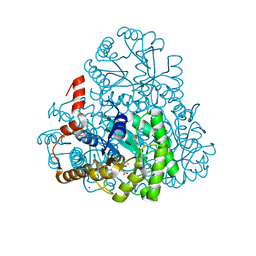 | | Structure of oxaloacetate acetylhydrolase in complex with the inhibitor 3,3-difluorooxalacetate | | Descriptor: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Herzberg, O, Chen, C. | | Deposit date: | 2010-03-03 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
J.Biol.Chem., 285, 2010
|
|
3OHI
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with 3-hydroxy-2-pyridone | | Descriptor: | ({3-hydroxy-2-oxo-4-[2-(phosphonooxy)ethyl]pyridin-1(2H)-yl}methyl)phosphonic acid, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Herzberg, O, Galkin, A. | | Deposit date: | 2010-08-17 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis and evaluation of first generation inhibitors of the Giardia lamblia fructose-1,6-biphosphate aldolase.
J.Inorg.Biochem., 105, 2010
|
|
3QYM
 
 | |
2DUA
 
 | |
1BLH
 
 | |
1BLC
 
 | |
6W4Q
 
 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J, Herzberg, O. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
1ALQ
 
 | |
4ZNB
 
 | | METALLO-BETA-LACTAMASE (C181S MUTANT) | | Descriptor: | METALLO-BETA-LACTAMASE, SODIUM ION, ZINC ION | | Authors: | Li, Z, Herzberg, O. | | Deposit date: | 1998-10-20 | | Release date: | 1999-06-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural consequences of the active site substitution Cys181 --> Ser in metallo-beta-lactamase from Bacteroides fragilis.
Protein Sci., 8, 1999
|
|
6NW9
 
 | | CRYSTAL STRUCTURE OF A TAILSPIKE PROTEIN 3 (TSP3, ORF212) FROM ESCHERICHIA COLI O157:H7 BACTERIOPHAGE CBA120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J.Y, Herzberg, O. | | Deposit date: | 2019-02-06 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3).
Sci Rep, 9, 2019
|
|
8SOT
 
 | |
2R82
 
 | | Pyruvate phosphate dikinase (PPDK) triple mutant R219E/E271R/S262D adapts a second conformational state | | Descriptor: | Pyruvate, phosphate dikinase, SULFATE ION | | Authors: | Lim, K, Read, R.J, Chen, C.C, Herzberg, O. | | Deposit date: | 2007-09-10 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Swiveling domain mechanism in pyruvate phosphate dikinase.
Biochemistry, 46, 2007
|
|
1DJC
 
 | |
