2APS
 
 | | CU/ZN SUPEROXIDE DISMUTASE FROM ACTINOBACILLUS PLEUROPNEUMONIAE | | Descriptor: | COPPER (II) ION, PROTEIN (CU,ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Forest, K.T, Langford, P.R, Kroll, J.S, Getzoff, E.D. | | Deposit date: | 1999-02-11 | | Release date: | 1999-02-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cu,Zn superoxide dismutase structure from a microbial pathogen establishes a class with a conserved dimer interface.
J.Mol.Biol., 296, 2000
|
|
2PIL
 
 | | Crystallographic Structure of Phosphorylated Pilin from Neisseria: Phosphoserine Sites Modify Type IV Pilus Surface Chemistry | | Descriptor: | HEPTANE-1,2,3-TRIOL, PLATINUM (II) ION, TYPE 4 PILIN, ... | | Authors: | Forest, K.T, Dunham, S.A, Koomey, M, Tainer, J.A. | | Deposit date: | 1998-03-02 | | Release date: | 1998-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic structure reveals phosphorylated pilin from Neisseria: phosphoserine sites modify type IV pilus surface chemistry and fibre morphology.
Mol.Microbiol., 31, 1999
|
|
1AY2
 
 | | STRUCTURE OF THE FIBER-FORMING PROTEIN PILIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | HEPTANE-1,2,3-TRIOL, PLATINUM (II) ION, TYPE 4 PILIN, ... | | Authors: | Forest, K.T, Parge, H.E, Tainer, J.A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-04-29 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fibre-forming protein pilin at 2.6 A resolution.
Nature, 378, 1995
|
|
6NQC
 
 | |
2GSZ
 
 | |
3S7O
 
 | | Crystal Structure of the Infrared Fluorescent D207H variant of Deinococcus Bacteriophytochrome chromophore binding domain at 1.24 angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, GLYCEROL | | Authors: | Forest, K.T, Auldridge, M.E, Satyshur, K.A, Anstrom, D.M. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein.
J.Biol.Chem., 287, 2012
|
|
3S7P
 
 | |
7SQX
 
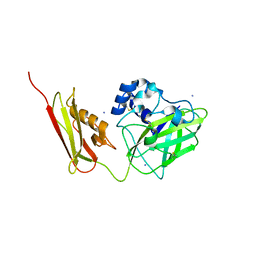 | | Crystal Structure of Pseudomonas aeruginosa lytic polysaccharide monooxygenase CbpD | | Descriptor: | AMMONIUM ION, Chitin-binding protein CbpD | | Authors: | Dade, C, Douzi, B, Ball, G, Voulhoux, R, Forest, K.T. | | Deposit date: | 2021-11-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of CbpD clarifies substrate-specificity motifs in chitin-active lytic polysaccharide monooxygenases.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4RWB
 
 | | Racemic influenza M2-TM crystallized from monoolein lipidic cubic phase | | Descriptor: | Matrix protein 2, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Mortenson, D.E, Steinkruger, J.D, Kreitler, D.F, Gellman, S.H, Forest, K.T. | | Deposit date: | 2014-12-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of a heterochiral coiled coil.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RWC
 
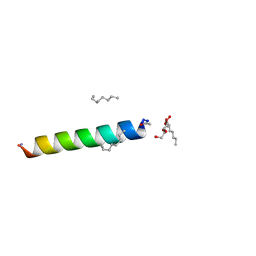 | | Racemic M2-TM crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Mortenson, D.E, Steinkruger, J.D, Kreitler, D.F, Gellman, S.H, Forest, K.T. | | Deposit date: | 2014-12-02 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High-resolution structures of a heterochiral coiled coil.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6VBS
 
 | |
6V7K
 
 | |
6VBT
 
 | |
4WMQ
 
 | |
4WN0
 
 | | Xenopus laevis embryonic epidermal lectin in complex with glycerol phosphate | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, PHOSPHATE ION, ... | | Authors: | Wangkanont, K, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2014-10-10 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Xenopus Embryonic Epidermal Lectin Reveal a Conserved Mechanism of Microbial Glycan Recognition.
J.Biol.Chem., 291, 2016
|
|
4WMY
 
 | |
4WMO
 
 | |
4WPB
 
 | | Vascular endothelial growth factor in complex with alpha/beta-VEGF-1 | | Descriptor: | Vascular endothelial growth factor A, alpha/beta-VEGF-1 | | Authors: | Kreitler, D.F, Checco, J.W, Gellman, S.H, Forest, K.T. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Targeting diverse protein-protein interaction interfaces with alpha / beta-peptides derived from the Z-domain scaffold.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6MPL
 
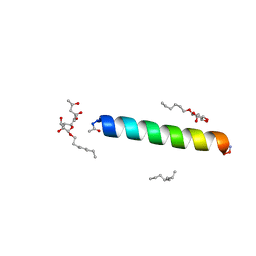 | | Racemic M2-TM I39A crystallized from racemic detergent | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6MPN
 
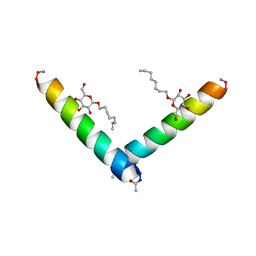 | | Racemic M2-TM I42E crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6MPM
 
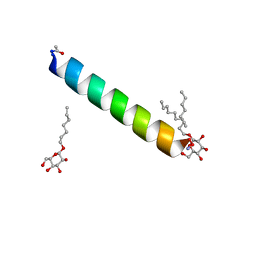 | | Racemic M2-TM I42A crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6NIV
 
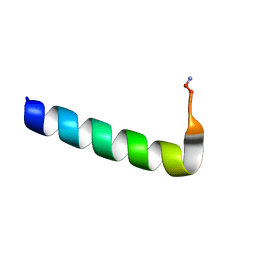 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
6O4M
 
 | | Racemic melittin | | Descriptor: | D-Melittin, Melittin, SULFATE ION | | Authors: | Kurgan, K.W, Bingman, C.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Retention of Native Quaternary Structure in Racemic Melittin Crystals.
J.Am.Chem.Soc., 141, 2019
|
|
2EWW
 
 | |
2EWV
 
 | |
