4RX6
 
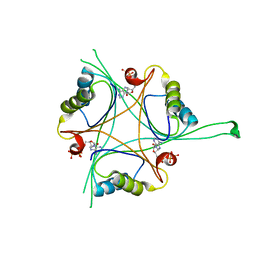 | | Structure of B. subtilis GlnK-ATP complex to 2.6 Angstrom | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory PII-like protein | | Authors: | Schumacher, M.A, Cuthbert, B, Tonthat, N, Chinnam, N.G, Whitfill, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5994 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4RS7
 
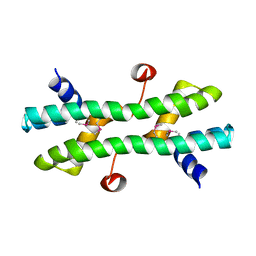 | |
4S0R
 
 | | Structure of GS-TnrA complex | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Chinnam, N.G, Cuthbert, B, Tonthat, N.K. | | Deposit date: | 2015-01-04 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4RS8
 
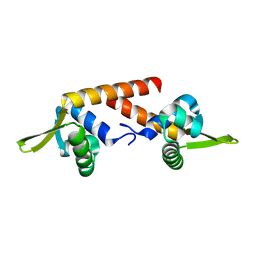 | |
1DH3
 
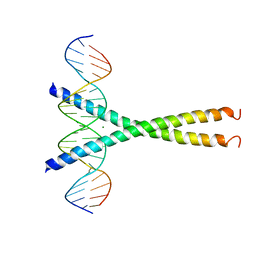 | |
8UFJ
 
 | | Structure of M. mazei GS(R167L-A168G) apo form | | Descriptor: | Glutamine synthetase, MAGNESIUM ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | M. mazei glutamine synthetase and glutamine synthetase-GlnK1 structures reveal enzyme regulation by oligomer modulation.
Nat Commun, 14, 2023
|
|
8SUA
 
 | | Structure of M. baixiangningiae DarR-ligand complex | | Descriptor: | 3-azanyl-3-(hydroxymethyl)-1,5,7,11-tetraoxa-6$l^{4}-boraspiro[5.5]undecan-9-ol, DarR | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SVA
 
 | | Structure of the Rhodococcus sp. USK13 DarR-20 bp DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*AP*CP*TP*CP*CP*GP*GP*AP*GP*TP*AP*TP*CP*TP*A)-3'), PHOSPHATE ION, TetR/AcrR family transcriptional regulator | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SUK
 
 | | Structure of Rhodococcus sp. USK13 DarR-c-di-AMP complex | | Descriptor: | DNA (5'-D(*AP*A)-3'), DarR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-12 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SV6
 
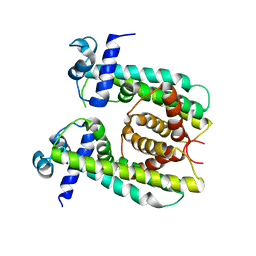 | | Structure of the M. smegmatis DarR protein | | Descriptor: | Fatty acid metabolism regulator protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8T5Y
 
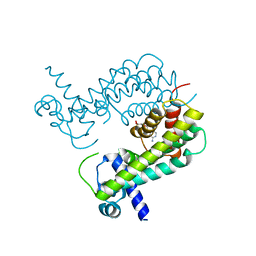 | |
8SVD
 
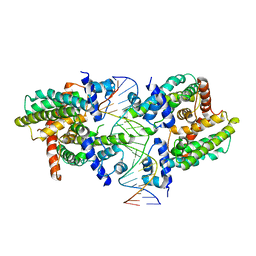 | |
8TFK
 
 | |
8TFC
 
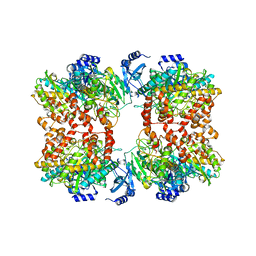 | |
8TGE
 
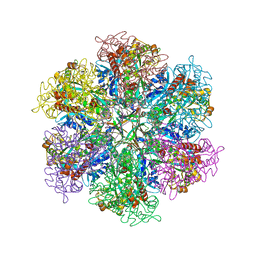 | |
8TFB
 
 | |
8TPK
 
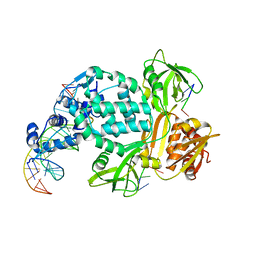 | | P6522 crystal form of C. crescentus DriD-ssDNA-DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*TP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*C)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site.
Nucleic Acids Res., 52, 2024
|
|
8TP8
 
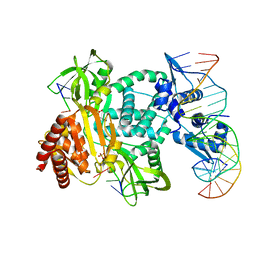 | | Structure of the C. crescentus WYL-activator, DriD, bound to ssDNA and cognate DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*TP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*C)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site.
Nucleic Acids Res., 52, 2024
|
|
2PUC
 
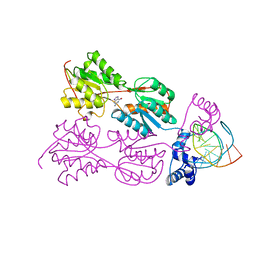 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
2PUD
 
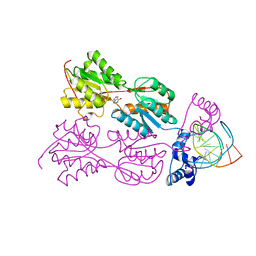 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
1DBQ
 
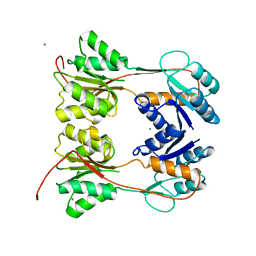 | | DNA-BINDING REGULATORY PROTEIN | | Descriptor: | MAGNESIUM ION, PURINE REPRESSOR | | Authors: | Schumacher, M.A, Choi, K.Y, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of corepressor-mediated specific DNA binding by the purine repressor.
Cell(Cambridge,Mass.), 83, 1995
|
|
1WET
 
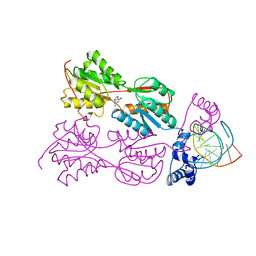 | | STRUCTURE OF THE PURR-GUANINE-PURF OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Glasfeld, A, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-04-27 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity.
J.Biol.Chem., 272, 1997
|
|
1HAB
 
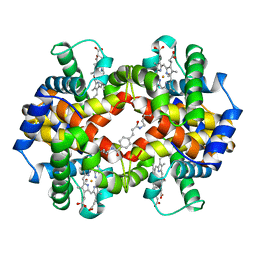 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 4-CARBOXYCINNAMIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1HAC
 
 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 2,6-DICARBOXYNAPHTHALENE, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
3TPE
 
 | | The phipa p3121 structure | | Descriptor: | Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
