3UD6
 
 | | Structural analyses of covalent enzyme-substrate analogue complexes reveal strengths and limitations of de novo enzyme design | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, RETRO-ALDOLASE, SULFATE ION | | Authors: | Baker, D, Stoddard, B.L, Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J, Hilvert, D. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
8T5E
 
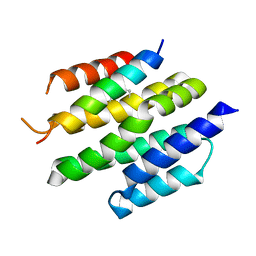 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Bcl-2-like protein 11, Bim_fulldiff | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8VC8
 
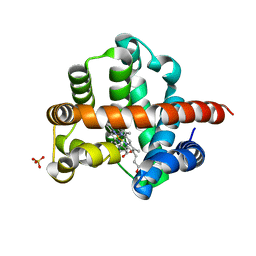 | |
8T5F
 
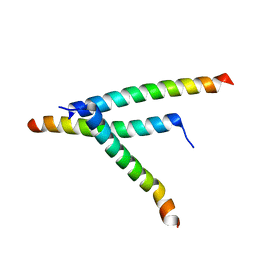 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Parathyroid hormone | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O0C
 
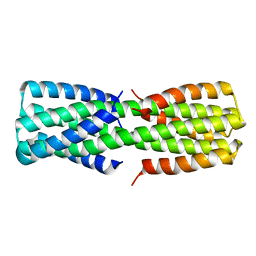 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8TCG
 
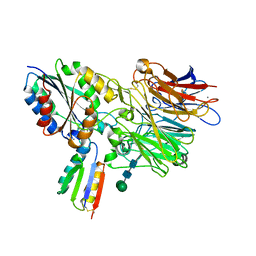 | | Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Integrin alpha-V heavy chain, ... | | Authors: | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | Deposit date: | 2023-06-30 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
8TCF
 
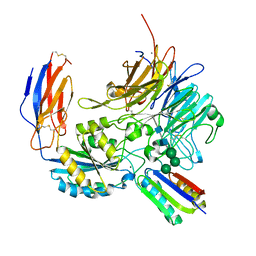 | | Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | Deposit date: | 2023-06-30 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
4UOT
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE 5H2L | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
4UOS
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
8GA7
 
 | |
8GA6
 
 | |
8G9J
 
 | |
8G9K
 
 | |
8UB3
 
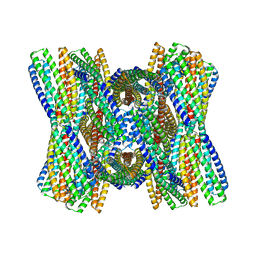 | | DpHF7 filament | | Descriptor: | DpHF7 filament | | Authors: | Lynch, E.M, Farrell, D, Shen, H, Kollman, J.M, DiMaio, F, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 2024
|
|
8UAO
 
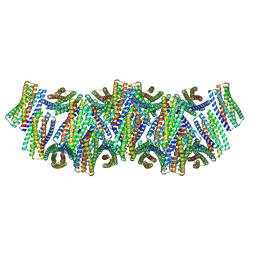 | | DpHF18 filament | | Descriptor: | DpHF18 | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-21 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 2024
|
|
4RJV
 
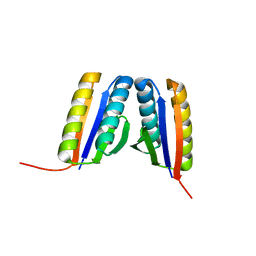 | | Crystal Structure of a De Novo Designed Ferredoxin Fold, Northeast Structural Genomics Consortium (NESG) Target OR461 | | Descriptor: | OR461 | | Authors: | O'Connell, P.T, Lin, Y.-R, Guan, R, Koga, N, Koga, R, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR461
To be published
|
|
4RZP
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR366. | | Descriptor: | Engineered Protein OR366 | | Authors: | Vorobiev, S, Parmeggiani, F, Dimaio, F, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal Structure of Engineered Protein OR366
To be Published
|
|
4RV1
 
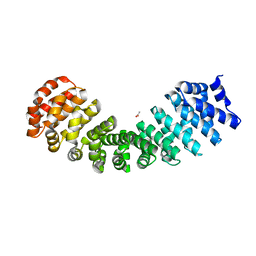 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR497. | | Descriptor: | ACETATE ION, Engineered Protein OR497 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Crystal Structure of Engineered Protein OR497.
To be Published
|
|
8UBG
 
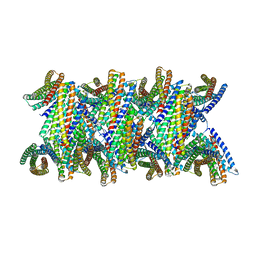 | | DpHF19 filament | | Descriptor: | DpHF19,Green fluorescent protein (Fragment) | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 2024
|
|
8E55
 
 | |
8E1E
 
 | |
8F53
 
 | |
8F54
 
 | |
8F4X
 
 | |
