1ANG
 
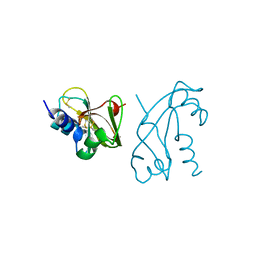 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN REVEALS THE STRUCTURAL BASIS FOR ITS FUNCTIONAL DIVERGENCE FROM RIBONUCLEASE | | Descriptor: | ANGIOGENIN | | Authors: | Acharya, K.R, Allen, S, Shapiro, R, Riordan, J.F, Vallee, B.L. | | Deposit date: | 1994-01-18 | | Release date: | 1995-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1ALC
 
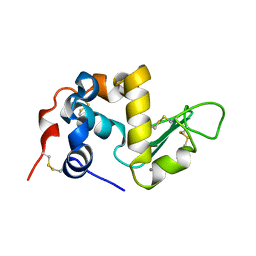 | |
1AGI
 
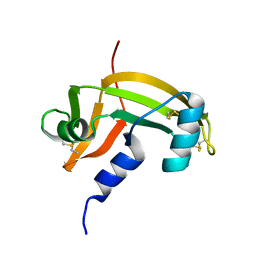 | |
1QIL
 
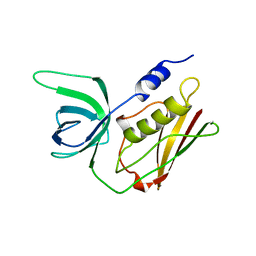 | |
6GPB
 
 | |
1STE
 
 | |
2QIL
 
 | |
1BBT
 
 | | METHODS USED IN THE STRUCTURE DETERMINATION OF FOOT AND MOUTH DISEASE VIRUS | | Descriptor: | FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP1), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP2), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP3), ... | | Authors: | Acharya, K.R, Fry, E.E, Logan, D.T, Stuart, D.I. | | Deposit date: | 1992-05-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Methods used in the structure determination of foot-and-mouth disease virus.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
1GPB
 
 | |
1LCL
 
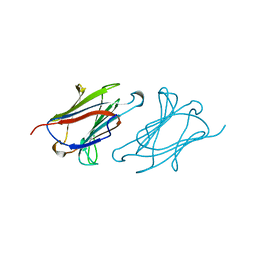 | | CHARCOT-LEYDEN CRYSTAL PROTEIN | | Descriptor: | LYSOPHOSPHOLIPASE | | Authors: | Acharya, K.R, Leonidas, D.D. | | Deposit date: | 1996-01-18 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Charcot-Leyden crystal protein, an eosinophil lysophospholipase, identifies it as a new member of the carbohydrate-binding family of galectins.
Structure, 3, 1995
|
|
3SEB
 
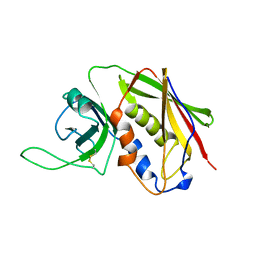 | | STAPHYLOCOCCAL ENTEROTOXIN B | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN B | | Authors: | Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1997-11-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of microbial superantigen staphylococcal enterotoxin B at 1.5 A resolution: implications for superantigen recognition by MHC class II molecules and T-cell receptors.
J.Mol.Biol., 277, 1998
|
|
5NJL
 
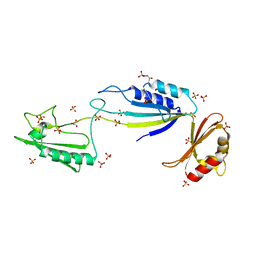 | | Cwp2 from Clostridium difficile | | Descriptor: | Cell surface protein (Putative S-layer protein), SULFATE ION | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-03-29 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cwp2 from Clostridium difficile exhibits an extended three domain fold and cell adhesion in vitro.
FEBS J., 284, 2017
|
|
5OQ3
 
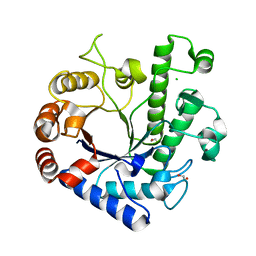 | | High resolution structure of the functional region of Cwp19 from Clostridium difficile | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cwp19, ... | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
FEBS J., 284, 2017
|
|
5OQ2
 
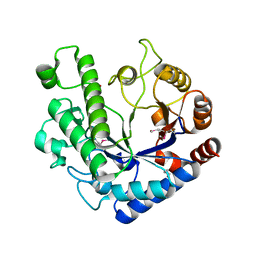 | | Se-SAD structure of the functional region of Cwp19 from Clostridium difficile | | Descriptor: | 1,2-ETHANEDIOL, Cwp19, PHOSPHATE ION | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-01 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
FEBS J., 284, 2017
|
|
7Q24
 
 | |
7Q26
 
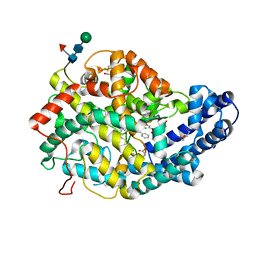 | | Crystal structure of Angiotensin-1 converting enzyme N-domain in complex with dual ACE/NEP inhibitor AD013 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-4-phenyl-butan-2-yl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2021-10-23 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the Requirements for Dual Angiotensin-Converting Enzyme C-Domain Selective/Neprilysin Inhibition.
J.Med.Chem., 65, 2022
|
|
7Q29
 
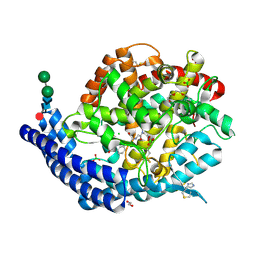 | | Crystal structure of Angiotensin-1 converting enzyme C-domain in complex with dual ACE/NEP inhibitor AD013 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-4-phenyl-butan-2-yl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2021-10-23 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Requirements for Dual Angiotensin-Converting Enzyme C-Domain Selective/Neprilysin Inhibition.
J.Med.Chem., 65, 2022
|
|
7Q25
 
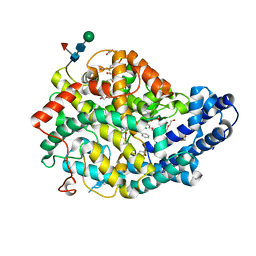 | |
7Q27
 
 | |
7Q28
 
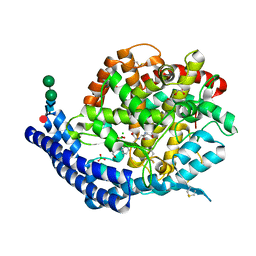 | |
7QPU
 
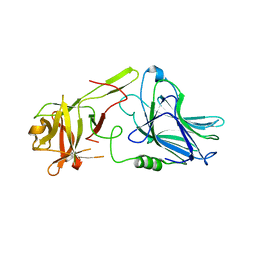 | | Botulinum neurotoxin A5 cell binding domain in complex with GM1b oligosaccharide | | Descriptor: | Botulinum neurotoxin sub-type A5, DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
7QPT
 
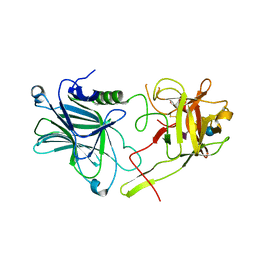 | | Botulinum neurotoxin A4 cell binding domain in complex with GD1a oligosaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type A | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mojanaga, O.O. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
4UW4
 
 | | Human galectin-7 in complex with a galactose based dendron D2-1. | | Descriptor: | DENDRON D2-1, GALECTIN-7 | | Authors: | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
4UW5
 
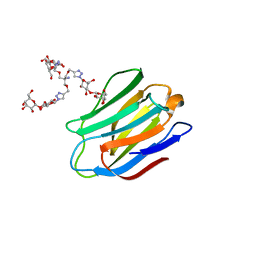 | | Human galectin-7 in complex with a galactose based dendron D2-2. | | Descriptor: | DENDRON D2-1, HUMAN GALECTIN-7 | | Authors: | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
4UW3
 
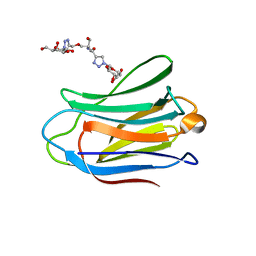 | | Human galectin-7 in complex with a galactose based dendron D1. | | Descriptor: | DENDRON-D1, HUMAN GALECTIN-7 | | Authors: | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
