7LFO
 
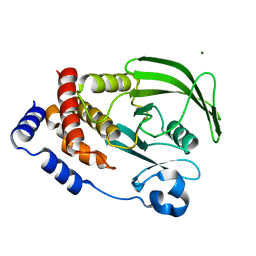 | | Protein Tyrosine Phosphatase 1B | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J.M. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
6W30
 
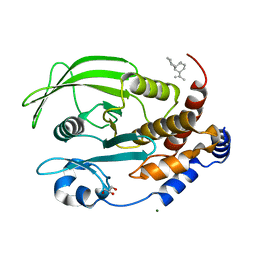 | | Protein Tyrosine Phosphatase 1B Bound to Amorphadiene | | Descriptor: | Amorphadiene, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J. | | Deposit date: | 2020-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
6B0N
 
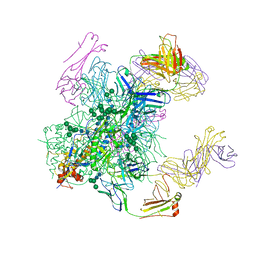 | | Crystal structure of the cleavage-independent prefusion HIV Env glycoprotein trimer of the clade A BG505 isolate (NFL construct) in complex with Fabs PGT122 and PGV19 at 3.39 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp140, ... | | Authors: | Sarkar, A, Irimia, A, Wilson, I.A. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer.
Nat Commun, 9, 2018
|
|
6AVN
 
 | |
8E1P
 
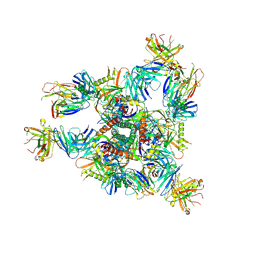 | | Crystal structure of BG505 SOSIP.v4.1-GT1.2 trimer in complex with gl-PGV20 and PGT124 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505-SOSIP.v4.1-GT1.2gp120, ... | | Authors: | Sarkar, A, Kumar, S, Wilson, I.A. | | Deposit date: | 2022-08-11 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.82 Å) | | Cite: | Germline-targeting HIV-1 Env vaccination induces VRC01-class antibodies with rare insertions.
Cell Rep Med, 4, 2023
|
|
5JOR
 
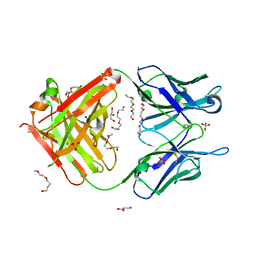 | | Crystal structure of unbound anti-glycan antibody Fab14.22 at 2.2 A | | Descriptor: | Fab 14.22 light chain, Fab14.22 heavy chain, GLYCEROL, ... | | Authors: | Sarkar, A, Irimia, A, Teyton, L, Wilson, I.A. | | Deposit date: | 2016-05-02 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | T cells control the generation of nanomolar-affinity anti-glycan antibodies.
J. Clin. Invest., 127, 2017
|
|
5JOP
 
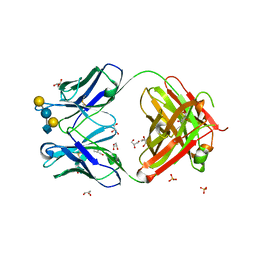 | | Crystal structure of anti-glycan antibody Fab14.22 in complex with Streptococcus pneumoniae serotype 14 tetrasaccharide at 1.75 A | | Descriptor: | Fab 14.22 light chain, Fab14.22 heavy chain, GLYCEROL, ... | | Authors: | Sarkar, A, Irimia, A, Teyton, L, Wilson, I.A. | | Deposit date: | 2016-05-02 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | T cells control the generation of nanomolar-affinity anti-glycan antibodies.
J. Clin. Invest., 127, 2017
|
|
5IF0
 
 | |
5IES
 
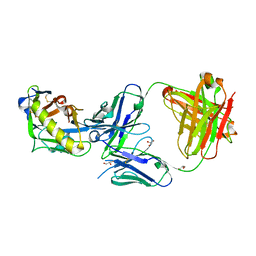 | |
5IFA
 
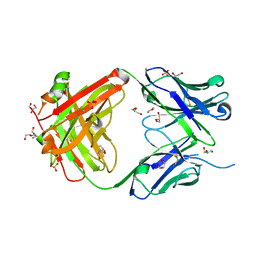 | |
6D2P
 
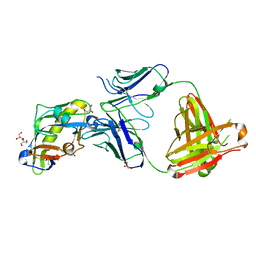 | |
6ME1
 
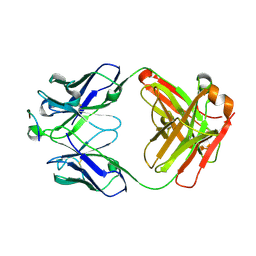 | |
6MCO
 
 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P23 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-08-31 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
6MDT
 
 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P63 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.816 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
3ZLL
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-azanyl-6-oxidanyl-1-(phenylmethyl)pyrimidine-2,4-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2013-02-01 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
3ZLK
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-AMINO-1-BENZYL-2,4-DIOXO-1,2,3,4-TETRAHYDROPYRIMIDIN-5-YL)BENZENESULFONAMIDE | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2013-02-01 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B42
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-27 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4ASY
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-05-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B2X
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-amino-1-butyl-5-(cyclopentylamino)pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B3U
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-amino-1-benzyl-5-(ethylamino)pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B4B
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 1-(2-phenylethyl)pyrimidine-2,4,6(1H,3H,5H)-trione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
6CE0
 
 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
4XC1
 
 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 GP41: crystals cryoprotected with sn-Glycerol 3-phosphate | | Descriptor: | 4E10 FAB HEAVY CHAIN, 4E10 FAB LIGHT CHAIN, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XCE
 
 | |
4XBE
 
 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41: crystals cryoprotected with sphingomyelin (02:0 SM (d18:1/2:0)). | | Descriptor: | 4E10 FAB LIGHT CHAIN, 4E10 Fab heavy chain, PEPTIDE FRAGMENT OF HIV GLYCOPROTEIN (GP41) including the region 671-683 of the MPER, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-16 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
