1H7I
 
 | |
1EA8
 
 | |
1BZ4
 
 | | APOLIPOPROTEIN E3 (APO-E3), TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
12E8
 
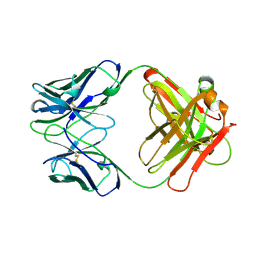 | | 2E8 FAB FRAGMENT | | Descriptor: | IGG1-KAPPA 2E8 FAB (HEAVY CHAIN), IGG1-KAPPA 2E8 FAB (LIGHT CHAIN) | | Authors: | Rupp, B, Trakhanov, S. | | Deposit date: | 1998-03-14 | | Release date: | 1998-08-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a monoclonal 2E8 Fab antibody fragment specific for the low-density lipoprotein-receptor binding region of apolipoprotein E refined at 1.9 A.
Acta Crystallogr.,Sect.D, null, 1999
|
|
1B68
 
 | |
6FAK
 
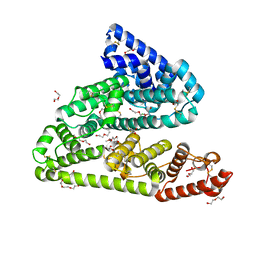 | | Human afamin orthorhombic crystal form by controlled hydration | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rupp, B, Naschberger, A, Bowler, M.W. | | Deposit date: | 2017-12-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Controlled dehydration, structural flexibility and gadolinium MRI contrast compound binding in the human plasma glycoprotein afamin.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5OKL
 
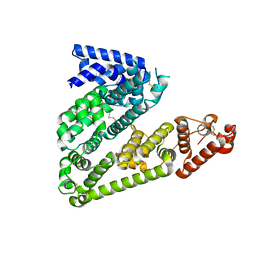 | | Human afamin monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Afamin, ... | | Authors: | Rupp, B, Naschberger, A, Bowler, M.W. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Evidence for a Role of the Multi-functional Human Glycoprotein Afamin in Wnt Transport.
Structure, 25, 2017
|
|
6RQ7
 
 | | Gadolinium MRI contrast compound binding in human plasma glycoprotein afamin - resurrection of highly anisotropic data | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rupp, B, Bowler, M.W, Naschberger, A, Juyoux, P, vonVelsen, J. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Controlled dehydration, structural flexibility and gadolinium MRI contrast compound binding in the human plasma glycoprotein afamin.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6SBI
 
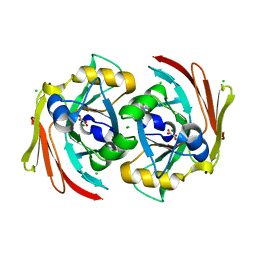 | | X-ray structure of murine Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) in complex with inhibitor oxalate | | Descriptor: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | Authors: | Rupp, B, Naschberger, A, Weiss, A.K.H. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
6SBJ
 
 | | X-ray structure of mus musculus Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) apo-form uuncomplexed | | Descriptor: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | Authors: | Rupp, B, Naschberger, A, Weiss, A.K.H. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
1NFN
 
 | | APOLIPOPROTEIN E3 (APOE3) | | Descriptor: | APOLIPOPROTEIN E3 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1NFO
 
 | | APOLIPOPROTEIN E2 (APOE2, D154A MUTATION) | | Descriptor: | APOLIPOPROTEIN E2 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1OR2
 
 | | APOLIPOPROTEIN E3 (APOE3) TRUNCATION MUTANT 165 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Rupp, B, Segelke, B.W, Forstner, M. | | Deposit date: | 1999-03-25 | | Release date: | 2000-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1OR3
 
 | | APOLIPOPROTEIN E3 (APOE3), TRIGONAL TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B.W. | | Deposit date: | 1998-12-01 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1UP5
 
 | | Chicken Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Wilson, M.A, Rupp, B. | | Deposit date: | 2003-09-27 | | Release date: | 2005-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization and Preliminary X-Ray Analysis of Two New Crystal Forms of Calmodulin
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5L88
 
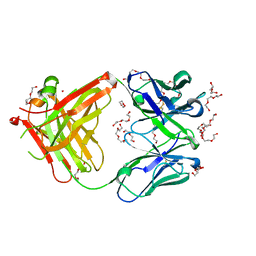 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSILATED, CRYSTAL FORM I, non-parsimonious model | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Anti-afamin antibody N14, Fab fragment, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5L9D
 
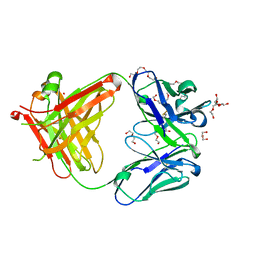 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSYLATED, CRYSTAL FORM I, parsimonious model | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-10 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5L7X
 
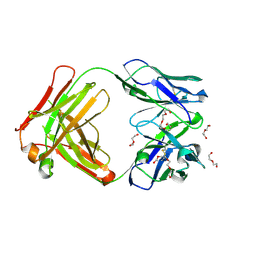 | | Afamin antibody fragment, N14 Fab, L1- glycosylated, crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Mouse Antibody Fab Fragment, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5LGH
 
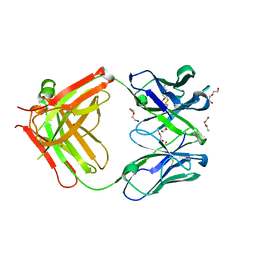 | | Afamin antibody fragment, N14 Fab, L1- glycosilated, crystal form II, same as 5L7X, but isomorphous setting indexed same as 5L88, 5L9D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, MOUSE ANTIBODY FAB FRAGMENT, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1A8D
 
 | | TETANUS TOXIN C FRAGMENT | | Descriptor: | GOLD ION, TETANUS NEUROTOXIN | | Authors: | Knapp, M, Rupp, B. | | Deposit date: | 1998-03-23 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The 1.61 Angstrom Structure of the Tetanus Toxin Ganglioside Binding Region: Solved by MAD and Mir Phase Combination
Am.Cryst.Assoc.,Abstr.Papers (Annual Meeting), 25, 1998
|
|
8QXQ
 
 | | PsiM in complex with SAH and psilocybin | | Descriptor: | CHLORIDE ION, Psilocybin synthase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-10-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
1BPI
 
 | |
7PGP
 
 | |
7PGQ
 
 | | GAP-SecPH region of human neurofibromin isoform 2 in closed conformation. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
2A2J
 
 | | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis | | Descriptor: | Pyridoxamine 5'-phosphate oxidase | | Authors: | Pedelacq, J.-D, Rho, B.-S, Kim, C.-Y, Waldo, G.S, Lekin, T.P, Segelke, B.W, Rupp, B, Hung, L.-W, Kim, S.-I, Terwilliger, T.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis.
Proteins, 62, 2005
|
|
