1FRO
 
 | |
1YCB
 
 | | DISTAL POCKET POLARITY IN LIGAND BINDING TO MYOGLOBIN: DEOXY AND CARBONMONOXY FORMS OF A THREONINE68 (E11) MUTANT INVESTIGATED BY X-RAY CRYSTALLOGRAPHY AND INFRARED SPECTROSCOPY | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cameron, A.D, Smerdon, S.J, Wilkinson, A.J, Habash, J, Helliwell, J.R. | | Deposit date: | 1993-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distal pocket polarity in ligand binding to myoglobin: deoxy and carbonmonoxy forms of a threonine68(E11) mutant investigated by X-ray crystallography and infrared spectroscopy.
Biochemistry, 32, 1993
|
|
1YCA
 
 | | DISTAL POCKET POLARITY IN LIGAND BINDING TO MYOGLOBIN: DEOXY AND CARBONMONOXY FORMS OF A THREONINE68 (E11) MUTANT INVESTIGATED BY X-RAY CRYSTALLOGRAPHY AND INFRARED SPECTROSCOPY | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cameron, A.D, Smerdon, S.J, Wilkinson, A.J, Habash, J, Helliwell, J.R. | | Deposit date: | 1993-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distal pocket polarity in ligand binding to myoglobin: deoxy and carbonmonoxy forms of a threonine68(E11) mutant investigated by X-ray crystallography and infrared spectroscopy.
Biochemistry, 32, 1993
|
|
1QIP
 
 | | HUMAN GLYOXALASE I COMPLEXED WITH S-P-NITROBENZYLOXYCARBONYLGLUTATHIONE | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (LACTOYLGLUTATHIONE LYASE), S-P-NITROBENZYLOXYCARBONYLGLUTATHIONE, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-06-14 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Reaction mechanism of glyoxalase I explored by an X-ray crystallographic analysis of the human enzyme in complex with a transition state analogue.
Biochemistry, 38, 1999
|
|
1QIN
 
 | | HUMAN GLYOXALASE I COMPLEXED WITH S-(N-HYDROXY-N-P-IODOPHENYLCARBAMOYL) GLUTATHIONE | | Descriptor: | PROTEIN (LACTOYLGLUTATHIONE LYASE), S-(N-HYDROXY-N-IODOPHENYLCARBAMOYL)GLUTATHIONE, ZINC ION | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-06-14 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of glyoxalase I explored by an X-ray crystallographic analysis of the human enzyme in complex with a transition state analogue.
Biochemistry, 38, 1999
|
|
1GSE
 
 | |
1QH5
 
 | | HUMAN GLYOXALASE II WITH S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE | | Descriptor: | GLUTATHIONE, PROTEIN (HYDROXYACYLGLUTATHIONE HYDROLASE), S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-05-11 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
Structure Fold.Des., 7, 1999
|
|
1QH3
 
 | | HUMAN GLYOXALASE II WITH CACODYLATE AND ACETATE IONS PRESENT IN THE ACTIVE SITE | | Descriptor: | ACETATE ION, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-05-10 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
Structure Fold.Des., 7, 1999
|
|
1BH5
 
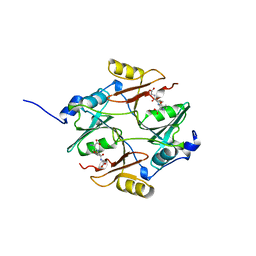 | | HUMAN GLYOXALASE I Q33E, E172Q DOUBLE MUTANT | | Descriptor: | LACTOYLGLUTATHIONE LYASE, S-HEXYLGLUTATHIONE, ZINC ION | | Authors: | Cameron, A.D, Jones, T.A. | | Deposit date: | 1998-06-13 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Involvement of an active-site Zn2+ ligand in the catalytic mechanism of human glyoxalase I.
J.Biol.Chem., 273, 1998
|
|
6H7B
 
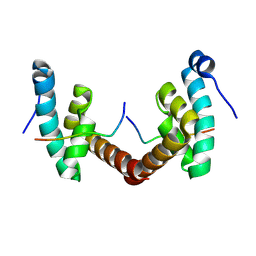 | | Structure of Leishmania PABP1 (domain J) complexed with a peptide containing the PAM2 motif of eIF4E4. | | Descriptor: | HIS-HIS-MET-ASN-PRO-ASN-ALA-THR-GLU-PHE-MET-PRO, Polyadenylate-binding protein | | Authors: | Cameron, A.D, Firczuk, H, dos Santos Rodrigues, F.H, McCarthy, J.E.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Leishmania PABP1-eIF4E4 interface: a novel 5'-3' interaction architecture for trans-spliced mRNAs.
Nucleic Acids Res., 47, 2019
|
|
6H7A
 
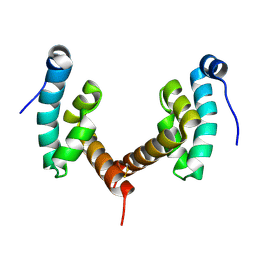 | |
2J50
 
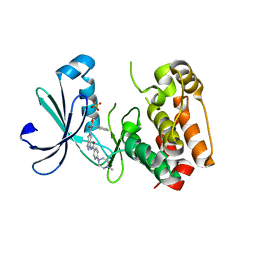 | | Structure of Aurora-2 in complex with PHA-739358 | | Descriptor: | N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6, SULFATE ION | | Authors: | Cameron, A.D, Izzo, G, Storici, P, Rusconi, L, Fancelli, D, Varasi, M, Berta, D, Bindi, S, Forte, B, Severino, D, Tonani, R, Vianello, P. | | Deposit date: | 2006-09-08 | | Release date: | 2006-11-06 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 1,4,5,6-Tetrahydropyrrolo[3,4-C]Pyrazoles: Identification of a Potent Aurora Kinase Inhibitor with a Favorable Antitumor Kinase Inhibition Profile.
J.Med.Chem., 49, 2006
|
|
2JGC
 
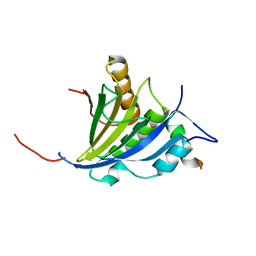 | | Structure of the human eIF4E homologous protein, 4EHP without ligand bound | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 2, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1 | | Authors: | Cameron, A.D, Rosettani, P, Knapp, S, Vismara, M.G, Rusconi, L. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms.
J. Mol. Biol., 368, 2007
|
|
2JGB
 
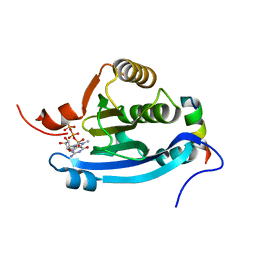 | | Structure of human eIF4E homologous protein 4EHP with m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 2, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1 | | Authors: | Cameron, A.D, Rosettani, P, Knapp, S, Vismara, M.G, Rusconi, L. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms.
J. Mol. Biol., 368, 2007
|
|
1GSD
 
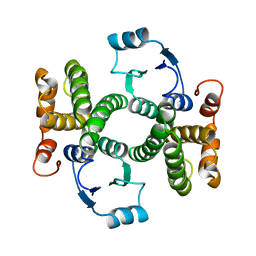 | |
1GSF
 
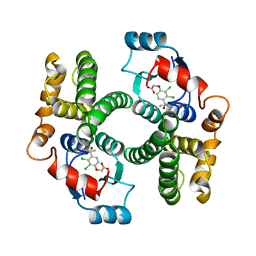 | | GLUTATHIONE TRANSFERASE A1-1 COMPLEXED WITH ETHACRYNIC ACID | | Descriptor: | ETHACRYNIC ACID, GLUTATHIONE TRANSFERASE A1-1 | | Authors: | L'Hermite, G, Sinning, I, Cameron, A.D, Jones, T.A. | | Deposit date: | 1995-06-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of human alpha-class glutathione transferase A1-1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate.
Structure, 3, 1995
|
|
8OYF
 
 | |
8OYG
 
 | |
7APE
 
 | | Crystal structure of LpqY from Mycobacterium thermoresistible in complex with trehalose | | Descriptor: | Lipoprotein (Sugar-binding) lpqY, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Furze, C.M, Guy, C.M, Angula, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter.
J.Biol.Chem., 296, 2021
|
|
5I6C
 
 | | The structure of the eukaryotic purine/H+ symporter, UapA, in complex with Xanthine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Uric acid-xanthine permease, XANTHINE | | Authors: | Alguel, Y, Amillis, S, Leung, J, Lambrinidis, G, Capaldi, S, Scull, N.J, Craven, G, Iwata, S, Armstrong, A, Mikros, E, Diallinas, G, Cameron, A.D, Byrne, B. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-27 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of eukaryotic purine/H(+) symporter UapA suggests a role for homodimerization in transport activity.
Nat Commun, 7, 2016
|
|
6R1B
 
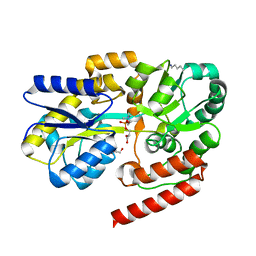 | | Crystal structure of UgpB from Mycobacterium tuberculosis in complex with glycerophosphocholine | | Descriptor: | 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fenn, J, Nepravishta, R, Guy, C.S, Harrison, J, Angulo, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27000213 Å) | | Cite: | Structural Basis of Glycerophosphodiester Recognition by theMycobacterium tuberculosisSubstrate-Binding Protein UgpB.
Acs Chem.Biol., 14, 2019
|
|
6SCT
 
 | | Cryo-EM structure of the consensus triskelion hub of the clathrin coat complex | | Descriptor: | Clathrin heavy chain, Clathrin light chain | | Authors: | Morris, K.L, Cameron, A.D, Sessions, R, Smith, C.J. | | Deposit date: | 2019-07-25 | | Release date: | 2019-10-02 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3VG9
 
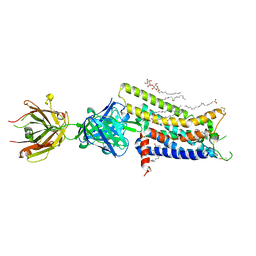 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 2.7 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
3VGA
 
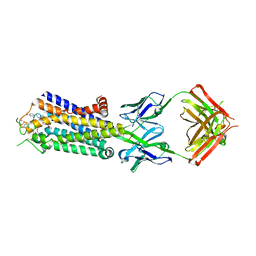 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 3.1 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, antibody fab fragment heavy chain, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
1RKS
 
 | |
