117E
 
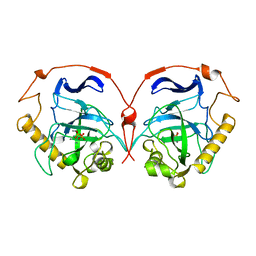 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
1AFT
 
 | | SMALL SUBUNIT C-TERMINAL INHIBITORY PEPTIDE OF MOUSE RIBONUCLEOTIDE REDUCTASE AS BOUND TO THE LARGE SUBUNIT, NMR, 26 STRUCTURES | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE | | Authors: | Laub, P.B, Fisher, A.L, Furst, G.T, Barwis, B.A, Hamann, C.S, Cooperman, B.S. | | Deposit date: | 1997-03-13 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an inhibitory R2 C-terminal peptide bound to mouse ribonucleotide reductase R1 subunit.
Nat.Struct.Biol., 2, 1995
|
|
1AIS
 
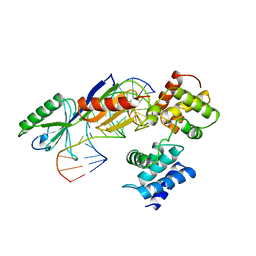 | | TATA-BINDING PROTEIN/TRANSCRIPTION FACTOR (II)B/TATA-BOX COMPLEX FROM PYROCOCCUS WOESEI | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*AP*CP*TP*TP*TP*(5IU)P*(5IU)P*AP*AP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*TP*TP*AP*AP*AP*AP*AP*GP*TP*AP*AP*GP*TP*T )-3'), PROTEIN (TATA-BINDING PROTEIN), ... | | Authors: | Kosa, P.F, Ghosh, G, Dedecker, B.S, Sigler, P.B. | | Deposit date: | 1997-04-24 | | Release date: | 1997-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AZQ
 
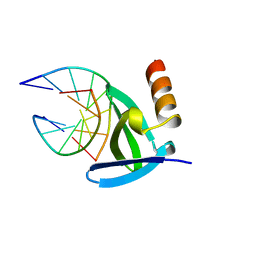 | | HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D BOUND WITH KINKED DNA DUPLEX | | Descriptor: | DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3'), PROTEIN (HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D) | | Authors: | Robinson, H, Gao, Y.-G, Mccrary, B.S, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Nature, 392, 1998
|
|
8UX7
 
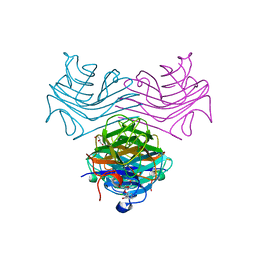 | | Dioclea megacarpa lectin (DmegA) complexed with X-Man | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Dioclea megacarpa lectin, ... | | Authors: | Oliveira, M.V, De Sloover, G, Osterne, V.J.S, Pinto-Junior, V.R, Sacramento-Neto, J.C, Van Damme, E.J.M, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2023-11-09 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dioclea megacarpa lectin (DmegA) complexed with X-Man
To Be Published
|
|
8SZO
 
 | | Canavalia villosa lectin in complex with alpha-methyl-mannoside | | Descriptor: | CALCIUM ION, Canavalia villosa lectin, GLYCEROL, ... | | Authors: | Cavada, B.S, Lossio, C.F, Pinto-Junior, V.R, Osterne, V.J.S, Oliveira, M.V, Neco, A.H.B, Nascimento, K.S. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lectin from Canavalia villosa seeds: A glucose/mannose-specific protein and a new tool for inflammation studies.
Int J Biol Macromol, 105, 2017
|
|
1AZP
 
 | | HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D BOUND WITH KINKED DNA DUPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3'), PROTEIN (HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D) | | Authors: | Robinson, H, Gao, Y.-G, Mccrary, B.S, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1997-11-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Nature, 392, 1998
|
|
1AZD
 
 | | CONCANAVALIN FROM CANAVALIA BRASILIENSIS | | Descriptor: | CALCIUM ION, CONBR, MANGANESE (II) ION | | Authors: | Sanz-Aparicio, J, Hermoso, J, Grangeiro, T.B, Calvete, J.J, Cavada, B.S. | | Deposit date: | 1997-11-16 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of Canavalia brasiliensis lectin suggests a correlation between its quaternary conformation and its distinct biological properties from Concanavalin A.
FEBS Lett., 405, 1997
|
|
1BNZ
 
 | | SSO7D HYPERTHERMOPHILE PROTEIN/DNA COMPLEX | | Descriptor: | 5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3', DNA-BINDING PROTEIN 7A | | Authors: | Gao, Y.-G, Su, S.-Y, Robinson, H, Padmanabhan, S, Lim, L, Mccrary, B.S, Edmondos, S.P, Shrive, J.W, Wang, A.H.-J. | | Deposit date: | 1998-07-31 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Nat.Struct.Biol., 5, 1998
|
|
8FFU
 
 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine, with the substrate bound | | Descriptor: | (2S,4S)-5-carbamimidamido-4-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pentanoic acid (non-preferred name), Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
8FFT
 
 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine | | Descriptor: | Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Lima, S.T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
4V4H
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with the antibiotic kasugamyin at 3.5A resolution. | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, ... | | Authors: | Schuwirth, B.S, Vila-Sanjurjo, A, Cate, J.H.D. | | Deposit date: | 2006-08-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structural analysis of kasugamycin inhibition of translation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5KAY
 
 | | Structure of Spelter bound to Zn2+ | | Descriptor: | SODIUM ION, Spelter, ZINC ION | | Authors: | Guffy, S.L, Der, B.S, Kuhlman, B. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-03 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the minimal determinants of zinc binding with computational protein design.
Protein Eng.Des.Sel., 29, 2016
|
|
4WCZ
 
 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
7N3K
 
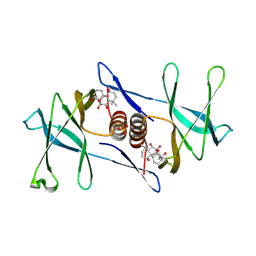 | | Oridonin-bound SARS-CoV-2 Nsp9 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A natural product compound inhibits coronaviral replication in vitro by binding to the conserved Nsp9 SARS-CoV-2 protein.
J.Biol.Chem., 297, 2021
|
|
4WGH
 
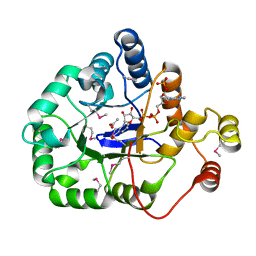 | | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution | | Descriptor: | ACETATE ION, Aldehyde reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Hillerich, B.S, Zimmerman, M.D, Chowdhury, S, Hammonds, J, Al Obaidi, N, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution
to be published
|
|
8PRK
 
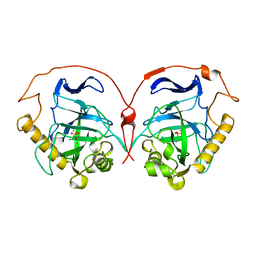 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-16 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
7YTN
 
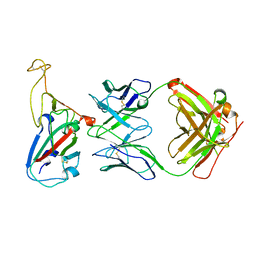 | |
7YTF
 
 | | Structure of OCPx2 from Nostoc flagelliforme CCNUN1 | | Descriptor: | Ketosteroid isomerase-related protein, beta,beta-carotene-4,4'-dione | | Authors: | Yang, Y.W, Chen, S.Z, Liu, K, Chen, M, Qiu, B.S. | | Deposit date: | 2022-08-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional specialization of expanded orange carotenoid protein paralogs in subaerial Nostoc species.
Plant Physiol., 192, 2023
|
|
7YTH
 
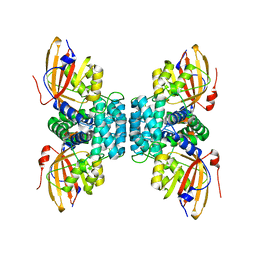 | | Structure of OCPx1 from Nostoc flagelliforme CCNUN1 | | Descriptor: | Ketosteroid isomerase-related protein | | Authors: | Yang, Y.W, Liu, K, Chen, S.Z, Chen, M, Qiu, B.S. | | Deposit date: | 2022-08-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Functional specialization of expanded orange carotenoid protein paralogs in subaerial Nostoc species.
Plant Physiol., 192, 2023
|
|
7ZCK
 
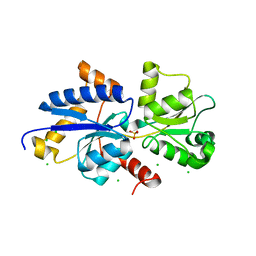 | | Room temperature crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Phosphonate ABC type transporter/ substrate binding component | | Authors: | Mikolajek, H, Shah, B.S, Paulsen, I.T, Sandy, J, Sanchez-Weatherby, J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
8B5X
 
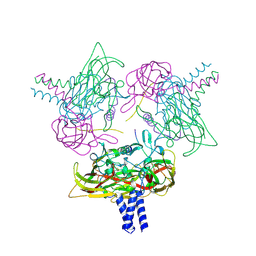 | | Crystal structure of the SUN1-KASH6 9:6 complex | | Descriptor: | CHLORIDE ION, Inositol 1,4,5-triphosphate receptor associated 2, POTASSIUM ION, ... | | Authors: | Erlandsen, B.S, Gurusaran, M, Davies, O.R. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of SUN1-KASH6 reveals an asymmetric LINC complex architecture compatible with nuclear membrane insertion.
Commun Biol, 7, 2024
|
|
8B46
 
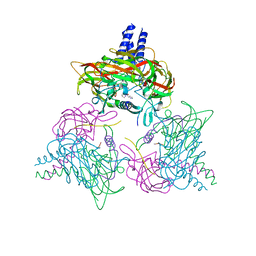 | | Crystal structure of the SUN1-KASH6 9:9 complex | | Descriptor: | CHLORIDE ION, Inositol 1,4,5-triphosphate receptor associated 2, POTASSIUM ION, ... | | Authors: | Gurusaran, M, Erlandsen, B.S, Davies, O.R. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The crystal structure of SUN1-KASH6 reveals an asymmetric LINC complex architecture compatible with nuclear membrane insertion.
Commun Biol, 7, 2024
|
|
8FKF
 
 | |
