1FJN
 
 | |
1D5Q
 
 | | SOLUTION STRUCTURE OF A MINI-PROTEIN REPRODUCING THE CORE OF THE CD4 SURFACE INTERACTING WITH THE HIV-1 ENVELOPE GLYCOPROTEIN | | Descriptor: | CHIMERIC MINI-PROTEIN | | Authors: | Vita, C, Drakopoulou, E, Vizzanova, J, Rochette, S, Martin, L, Menez, A, Roumestand, C, Yang, Y.S, Ylisastigui, L, Benjouad, A, Gluckman, J.C. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2OSG
 
 | | Solution Structure and Binding Property of the Domain-swapped Dimer of ZO2PDZ2 | | Descriptor: | Tight junction protein ZO-2 | | Authors: | Wu, J.W, Yang, Y.S, Zhang, J.H, Ji, P, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2007-02-05 | | Release date: | 2007-09-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Domain-swapped Dimerization of the Second PDZ Domain of ZO2 May Provide a Structural Basis for the Polymerization of Claudins
J.Biol.Chem., 282, 2007
|
|
4H00
 
 | | The crystal structure of mon-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4GZ7
 
 | | The crystal structure of Apo-dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, dihydropyrimidinase | | Authors: | Hsien, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structure Requirements for Post-translational Modification
To be Published
|
|
4H01
 
 | | The crystal structure of di-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4LCQ
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBI | | Descriptor: | (2S)-3-(carbamoylamino)-2-methylpropanoic acid, ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4LCS
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with hydantoin | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, ZINC ION, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S. | | Deposit date: | 2013-06-23 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4LCR
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBA | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
1KOC
 
 | | RNA APTAMER COMPLEXED WITH ARGININE, NMR | | Descriptor: | ARGININE, RNA (5'-R(P*AP*CP*AP*GP*GP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*CP*GP*U)-3') | | Authors: | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-08-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
1KOD
 
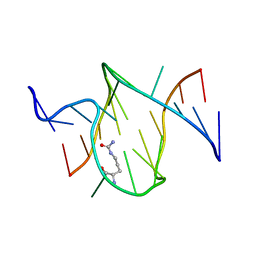 | | RNA APTAMER COMPLEXED WITH CITRULLINE, NMR | | Descriptor: | CITRULLINE, RNA (5'-R(P*AP*CP*GP*GP*UP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*UP*GP*U)-3') | | Authors: | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-11-08 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
2LCU
 
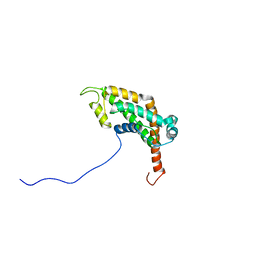 | | NMR structure of BC28.1 | | Descriptor: | Bc28.1 | | Authors: | Roumestand, C, Delbecq, S, Yang, Y. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2012-04-04 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Bc28.1, Major Erythrocyte-binding Protein from Babesia canis Merozoite Surface.
J.Biol.Chem., 287, 2012
|
|
2MCF
 
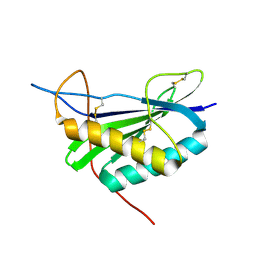 | | NMR structure of TGAM_1934 | | Descriptor: | TGAM_1934 | | Authors: | Yang, Y, Montet de Guillen, K, Roumestand, C. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Prioritizing targets for structural biology through the lens of proteomics: the archaeal protein TGAM_1934 from Thermococcus gammatolerans.
Proteomics, 15, 2015
|
|
1QTT
 
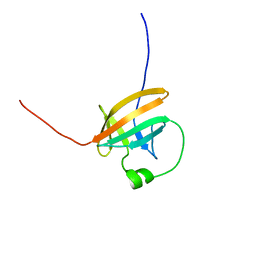 | | SOLUTION STRUCTURE OF THE ONCOPROTEIN P13MTCP1 | | Descriptor: | PRODUCT OF THE MTCP1 ONCOGENE | | Authors: | Guignard, L, Padilla, A, Mispelter, J, Yang, Y.-S, Stern, M.-H, Lhoste, J.-M, Roumestand, C. | | Deposit date: | 1999-06-29 | | Release date: | 2001-01-19 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and solution structure refinement of the 15N-labeled human oncogenic protein p13MTCP1: comparison with X-ray data.
J.Biomol.NMR, 17, 2000
|
|
1QTU
 
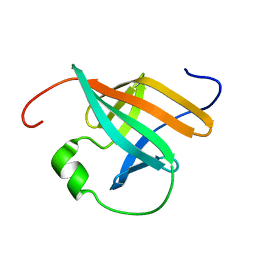 | | SOLUTION STRUCTURE OF THE ONCOPROTEIN P13MTCP1 | | Descriptor: | PROTEIN (PRODUCT OF THE MTCP1 ONCOGENE) | | Authors: | Guignard, L, Padilla, A, Mispelter, J, Yang, Y.-S, Stern, M.-H, Lhoste, J.-M, Roumestand, C. | | Deposit date: | 1999-06-29 | | Release date: | 2001-01-19 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and solution structure refinement of the 15N-labeled human oncogenic protein p13MTCP1: comparison with X-ray data.
J.Biomol.NMR, 17, 2000
|
|
1JSG
 
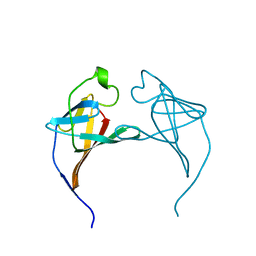 | | CRYSTAL STRUCTURE OF P14TCL1, AN ONCOGENE PRODUCT INVOLVED IN T-CELL PROLYMPHOCYTIC LEUKEMIA, REVEALS A NOVEL B-BARREL TOPOLOGY | | Descriptor: | ONCOGENE PRODUCT P14TCL1 | | Authors: | Hoh, F, Yang, Y.-S, Guignard, L, Padilla, A, Stern, R.-H, Lhoste, J.-M, Van Tilbeurgh, H. | | Deposit date: | 1997-12-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of p14TCL1, an oncogene product involved in T-cell prolymphocytic leukemia, reveals a novel beta-barrel topology.
Structure, 6, 1998
|
|
2QP4
 
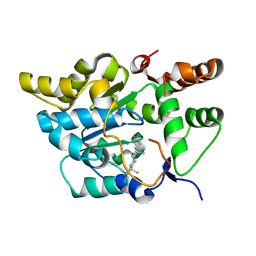 | |
2QP3
 
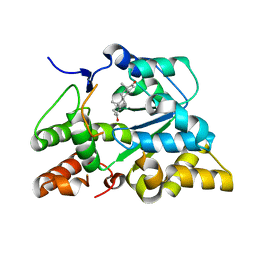 | |
1UXD
 
 | | Fructose repressor DNA-binding domain, NMR, 34 structures | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-01 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1UXC
 
 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-21 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
8D0E
 
 | | Human SARM1 TIR domain bound to NB-7 | | Descriptor: | 3-(4-chlorophenyl)-N-[4-methyl-3-(pyridin-4-yl)-1H-pyrazol-5-yl]propanamide, NAD(+) hydrolase SARM1 | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0J
 
 | | Apo Human SARM1 TIR domain | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0I
 
 | | Human SARM1 bound to an NB-3 eADPR adduct | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-imidazo[2,1-f]purin-3-yl-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0M
 
 | | Human CD38 ectodomain bound to a 78c-ADPR adduct | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[5-[4-[[4-(2-methoxyethoxy)cyclohexyl]amino]-1-methyl-2-oxidanylidene-quinolin-6-yl]-1,3-thiazol-3-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Gu, W. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0H
 
 | | Human SARM1 TIR domain bound to NB-3-GDPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
