1HST
 
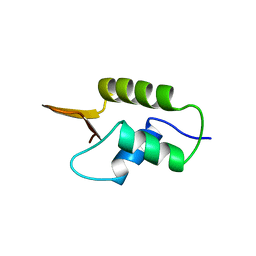 | | CRYSTAL STRUCTURE OF GLOBULAR DOMAIN OF HISTONE H5 AND ITS IMPLICATIONS FOR NUCLEOSOME BINDING | | Descriptor: | HISTONE H5 | | Authors: | Ramakrishnan, V, Finch, J.T, Graziano, V, Lee, P.L, Sweet, R.M. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of globular domain of histone H5 and its implications for nucleosome binding.
Nature, 362, 1993
|
|
1PKP
 
 | |
1BOB
 
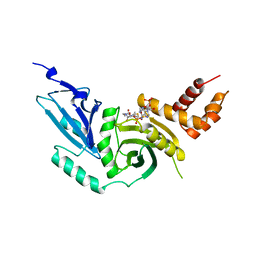 | | HISTONE ACETYLTRANSFERASE HAT1 FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH ACETYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, HISTONE ACETYLTRANSFERASE | | Authors: | Dutnall, R.N, Tafrov, S.T, Sternglanz, R, Ramakrishnan, V. | | Deposit date: | 1998-07-02 | | Release date: | 1999-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily.
Cell(Cambridge,Mass.), 94, 1998
|
|
8OZ0
 
 | | Structure of a human 48S translation initiation complex with eIF4F and eIF4A | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Fraser, C.S, Ramakrishnan, V. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6QG1
 
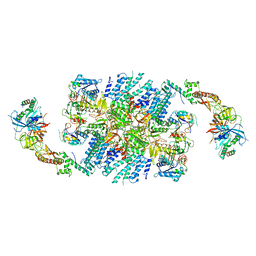 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model 2) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
7PD3
 
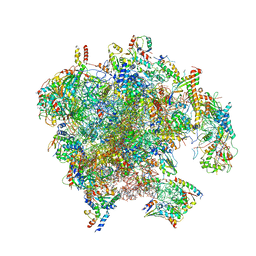 | | Structure of the human mitoribosomal large subunit in complex with NSUN4.MTERF4.GTPBP7 and MALSU1.L0R8F8.mt-ACP | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Chandrasekaran, V, Desai, N, Burton, N.O, Yang, H, Price, J, Miska, E.A, Ramakrishnan, V. | | Deposit date: | 2021-08-04 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualizing formation of the active site in the mitochondrial ribosome.
Elife, 10, 2021
|
|
4V5G
 
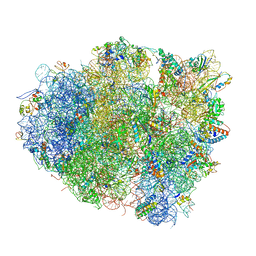 | | The crystal structure of the 70S ribosome bound to EF-Tu and tRNA | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2009-09-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA.
Science, 326, 2009
|
|
4V5Q
 
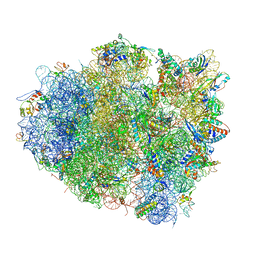 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near- cognate codon on the 70S ribosome | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3OTO
 
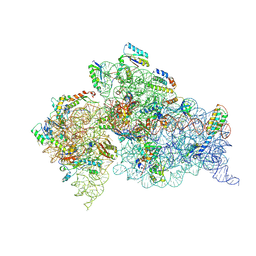 | | Crystal Structure of the 30S ribosomal subunit from a KsgA mutant of Thermus thermophilus (HB8) | | Descriptor: | 16S rRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demirci, H, Murphy IV, F, Belardinelli, R, Kelley, A.C, Ramakrishnan, V, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function.
Rna, 16, 2010
|
|
1A32
 
 | |
4WT8
 
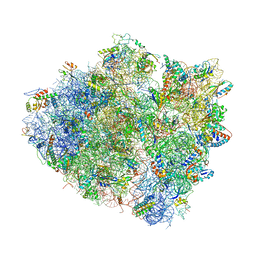 | | Crystal Structure of bactobolin A bound to 70S ribosome-tRNA complex | | Descriptor: | 23S rRNA (2899-MER), 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Amunts, A, Fiedorczuk, K, Ramakrishnan, V. | | Deposit date: | 2014-10-29 | | Release date: | 2015-01-21 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bactobolin A Binds to a Site on the 70S Ribosome Distinct from Previously Seen Antibiotics.
J.Mol.Biol., 427, 2015
|
|
5OOM
 
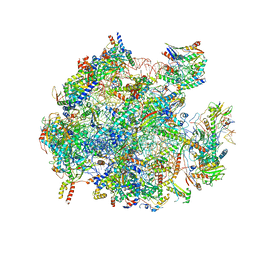 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OOL
 
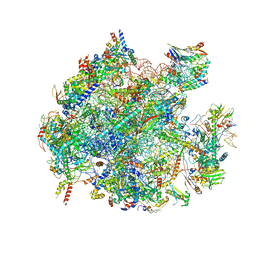 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5IQR
 
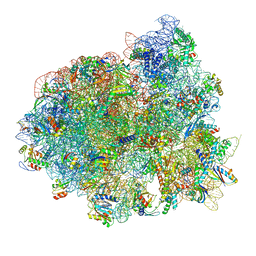 | | Structure of RelA bound to the 70S ribosome | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Brown, A, Fernandez, I.S, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome-dependent activation of stringent control.
Nature, 534, 2016
|
|
5IT9
 
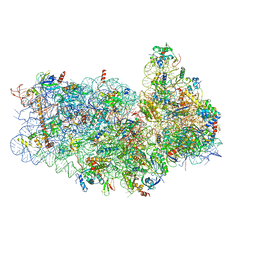 | | Structure of the yeast Kluyveromyces lactis small ribosomal subunit in complex with the cricket paralysis virus IRES. | | Descriptor: | 18S ribosomal RNA, Cricket paralysis virus IRES RNA, MAGNESIUM ION, ... | | Authors: | Murray, J, Savva, C.G, Shin, B.S, Dever, T.E, Ramakrishnan, V, Fernandez, I.S. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural characterization of ribosome recruitment and translocation by type IV IRES.
Elife, 5, 2016
|
|
5IT7
 
 | | Structure of the Kluyveromyces lactis 80S ribosome in complex with the cricket paralysis virus IRES and eEF2 | | Descriptor: | (2S)-1-amino-N,N,N-trimethyl-1-oxobutan-2-aminium, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Murray, J, Savva, C.G, Shin, B.S, Dever, T.E, Ramakrishnan, V, Fernandez, I.S. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural characterization of ribosome recruitment and translocation by type IV IRES.
Elife, 5, 2016
|
|
1FJG
 
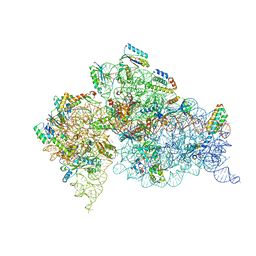 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
1TIF
 
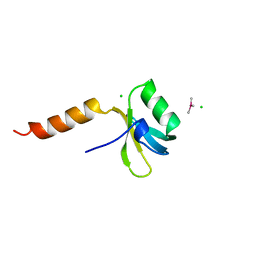 | | TRANSLATION INITIATION FACTOR 3 N-TERMINAL DOMAIN | | Descriptor: | CHLORIDE ION, TRANSLATION INITIATION FACTOR 3, TRIMETHYL LEAD ION | | Authors: | Biou, V, Shu, F, Ramakrishnan, V. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix.
EMBO J., 14, 1995
|
|
1TIG
 
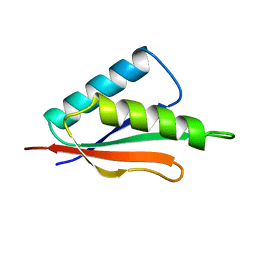 | | TRANSLATION INITIATION FACTOR 3 C-TERMINAL DOMAIN | | Descriptor: | TRANSLATION INITIATION FACTOR 3 | | Authors: | Biou, V, Shu, F, Ramakrishnan, V. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix.
EMBO J., 14, 1995
|
|
4V5K
 
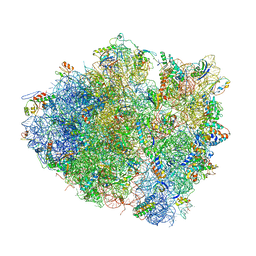 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1GHC
 
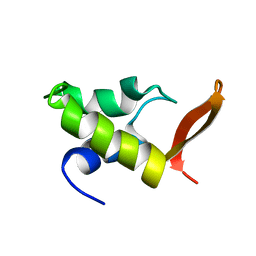 | | HOMO-AND HETERONUCLEAR TWO-DIMENSIONAL NMR STUDIES OF THE GLOBULAR DOMAIN OF HISTONE H1: FULL ASSIGNMENT, TERTIARY STRUCTURE, AND COMPARISON WITH THE GLOBULAR DOMAIN OF HISTONE H5 | | Descriptor: | GH1 | | Authors: | Cerf, C, Lippens, G, Ramakrishnan, V, Muyldermans, S, Segers, A, Wyns, L, Wodak, S.J, Hallenga, K. | | Deposit date: | 1994-05-16 | | Release date: | 1994-08-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5.
Biochemistry, 33, 1994
|
|
1QD7
 
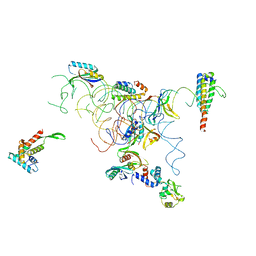 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
1HNW
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH TETRACYCLINE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HR0
 
 | | CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Morgan-Warren, R.J, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-20 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an initiation factor bound to the 30S ribosomal subunit.
Science, 291, 2001
|
|
1HNX
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
