1AMU
 
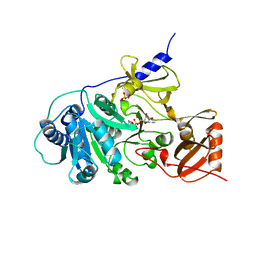 | | PHENYLALANINE ACTIVATING DOMAIN OF GRAMICIDIN SYNTHETASE 1 IN A COMPLEX WITH AMP AND PHENYLALANINE | | Descriptor: | ADENOSINE MONOPHOSPHATE, GRAMICIDIN SYNTHETASE 1, MAGNESIUM ION, ... | | Authors: | Conti, E, Stachelhaus, T, Marahiel, M.A, Brick, P. | | Deposit date: | 1997-06-18 | | Release date: | 1998-07-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S.
EMBO J., 16, 1997
|
|
6TA8
 
 | |
1DNY
 
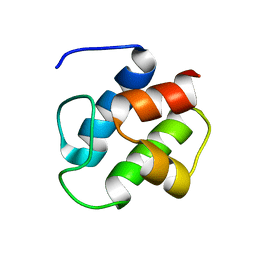 | | SOLUTION STRUCTURE OF PCP, A PROTOTYPE FOR THE PEPTIDYL CARRIER DOMAINS OF MODULAR PEPTIDE SYNTHETASES | | Descriptor: | NON-RIBOSOMAL PEPTIDE SYNTHETASE PEPTIDYL CARRIER PROTEIN | | Authors: | Weber, T, Baumgartner, R, Renner, C, Marahiel, M.A, Holak, T.A. | | Deposit date: | 1999-12-17 | | Release date: | 2000-05-17 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases.
Structure Fold.Des., 8, 2000
|
|
5OQZ
 
 | |
3E7W
 
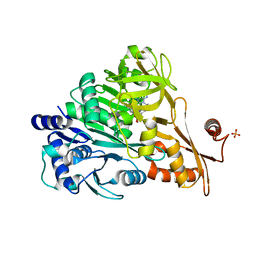 | | Crystal structure of DLTA: Implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1, PHOSPHATE ION | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
3E7X
 
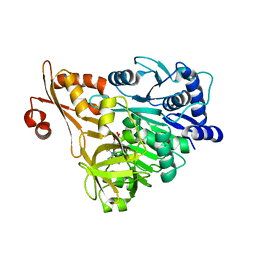 | | Crystal structure of DLTA: implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1 | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
2CBG
 
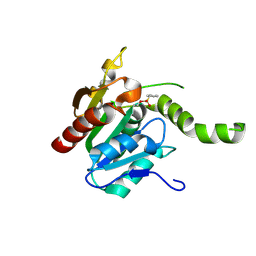 | |
2CB9
 
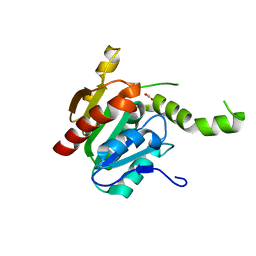 | |
2GDY
 
 | | Solution structure of the B. brevis TycC3-PCP in A-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
4MRT
 
 | | Structure of the Phosphopantetheine Transferase Sfp in Complex with Coenzyme A and a Peptidyl Carrier Protein | | Descriptor: | 4'-phosphopantetheinyl transferase sfp, COENZYME A, GLYCEROL, ... | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
4NAG
 
 | | Xanthomonins I III are a New Class of Lasso Peptides Featuringa Seven-Membered Macrolactam Ring | | Descriptor: | HEXANE-1,6-DIOL, Xanthomonin I | | Authors: | Hegemann, J.D, Zimmermann, M, Zhu, S, Steuber, H, Harms, K, Xie, X, Marahiel, M.A. | | Deposit date: | 2013-10-22 | | Release date: | 2014-04-30 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Xanthomonins I-III: A New Class of Lasso Peptides with a Seven-Residue Macrolactam Ring.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5D9E
 
 | | Crystal Structure of the Proline-rich Lasso Peptide Caulosegnin II | | Descriptor: | CHLORIDE ION, Caulosegnin II | | Authors: | Fage, C.D, Hegemann, J.D, Harms, K, Marahiel, M.A. | | Deposit date: | 2015-08-18 | | Release date: | 2016-02-17 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (0.859 Å) | | Cite: | The ring residue proline 8 is crucial for the thermal stability of the lasso peptide caulosegnin II.
Mol Biosyst, 12, 2016
|
|
5JRK
 
 | | Crystal Structure of the Sphingopyxin I Lasso Peptide Isopeptidase SpI-IsoP (SeMet-derived) | | Descriptor: | Dipeptidyl aminopeptidases/acylaminoacyl-peptidases-like protein, beta-D-glucopyranose | | Authors: | Fage, C.D, Hegemann, J.D, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5JRL
 
 | |
5JQF
 
 | | Crystal structure of the lasso peptide Sphingopyxin I (SpI) | | Descriptor: | Sphingopyxin I | | Authors: | Fage, C.D, Hegemann, J.D, Harms, K, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-04 | | Release date: | 2016-09-14 | | Last modified: | 2021-06-16 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
2RON
 
 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2016-10-26 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
1JMK
 
 | | Structural Basis for the Cyclization of the Lipopeptide Antibiotic Surfactin by the Thioesterase Domain SrfTE | | Descriptor: | SULFATE ION, Surfactin Synthetase | | Authors: | Bruner, S.D, Weber, T, Kohli, R.M, Schwarzer, D, Marahiel, M.A, Walsh, C.T, Stubbs, M.T. | | Deposit date: | 2001-07-18 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE.
Structure, 10, 2002
|
|
1MD9
 
 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB AND AMP | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MDF
 
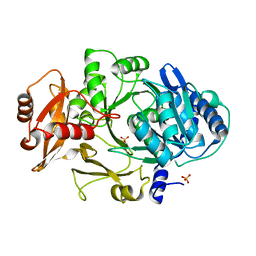 | | CRYSTAL STRUCTURE OF DhbE IN ABSENCE OF SUBSTRATE | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase, SULFATE ION | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MDB
 
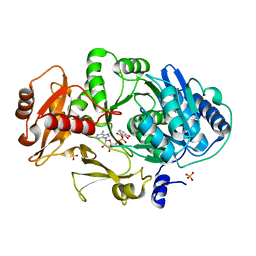 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB-ADENYLATE | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE, ... | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2WBQ
 
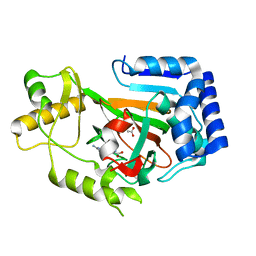 | | Crystal structure of VioC in complex with (2S,3S)-hydroxyarginine | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, ACETATE ION, L-ARGININE BETA-HYDROXYLASE | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
2WBP
 
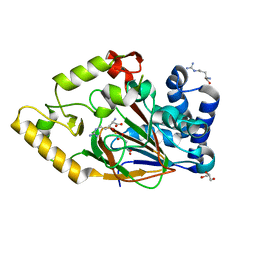 | | Crystal structure of VioC in complex with Fe(II), (2S,3S)- hydroxyarginine, and succinate | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, ARGININE, FE (II) ION, ... | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
2WBO
 
 | | Crystal structure of VioC in complex with L-arginine | | Descriptor: | ARGININE, FE (II) ION, L(+)-TARTARIC ACID, ... | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
2VSQ
 
 | | Structure of surfactin A synthetase C (SrfA-C), a nonribosomal peptide synthetase termination module | | Descriptor: | LEUCINE, SULFATE ION, SURFACTIN SYNTHETASE SUBUNIT 3 | | Authors: | Tanovic, A, Samel, S.A, Essen, L.-O, Marahiel, M.A. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the termination module of a nonribosomal peptide synthetase.
Science, 321, 2008
|
|
2WI8
 
 | | Crystal structure of the triscatecholate siderophore binding protein FeuA from Bacillus subtilis | | Descriptor: | CHLORIDE ION, IRON-UPTAKE SYSTEM-BINDING PROTEIN | | Authors: | Peuckert, F, Miethke, M, Albrecht, A.G, Essen, L.-O, Marahiel, M.A. | | Deposit date: | 2009-05-08 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis and Stereochemistry of Triscatecholate Siderophore Binding by Feua.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
