5LGM
 
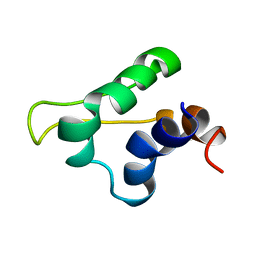 | | Gp5.7 mutant L42A | | Descriptor: | Fusion protein 5.5/5.7 | | Authors: | Liu, B, Matthews, S. | | Deposit date: | 2016-07-07 | | Release date: | 2017-08-16 | | Method: | SOLUTION NMR | | Cite: | Gp5.7 mutant L42A
to be published
|
|
4S20
 
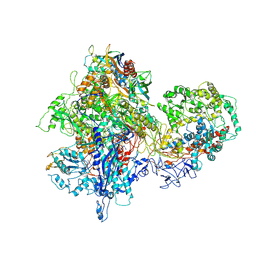 | | Structural basis for transcription reactivation by RapA | | Descriptor: | 5'-D(P*AP*CP*GP*AP*CP*TP*GP*AP*GP*CP*CP*GP*AP*TP*G)-3', 5'-R(P*AP*UP*CP*GP*GP*CP*UP*CP*A)-3', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Structural basis for transcription reactivation by RapA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6XQB
 
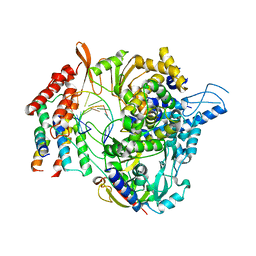 | | SARS-CoV-2 RdRp/RNA complex | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
7PEG
 
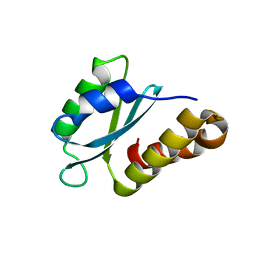 | | Structure of the sporulation/germination protein YhcN from Bacillus subtilis | | Descriptor: | Probable spore germination lipoprotein YhcN | | Authors: | Liu, B, Chan, H, Bauda, E, Contreras-Martel, C, Bellard, L, Villard, A.M, Mas, C, Neumann, E, Fenel, D, Favier, A, Serrano, M, Henriques, A.O.H, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into ring-building motif domains involved in bacterial sporulation.
J.Struct.Biol., 214, 2022
|
|
5TBZ
 
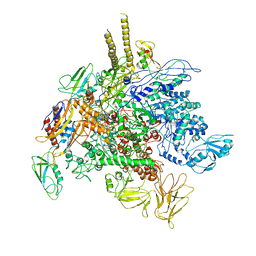 | | E. Coli RNA Polymerase complexed with NusG | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Steitz, T.A. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Structural insights into NusG regulating transcription elongation.
Nucleic Acids Res., 45, 2017
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
1UDE
 
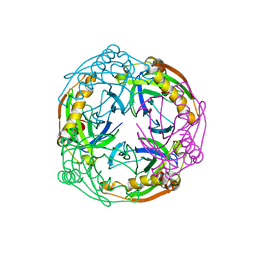 | | Crystal structure of the Inorganic pyrophosphatase from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Liu, B, Gao, R, Zhou, W, Bartlam, M, Rao, Z. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of the hyperthermophilic inorganic pyrophosphatase from the archaeon Pyrococcus horikoshii.
Biophys.J., 86, 2004
|
|
6GUM
 
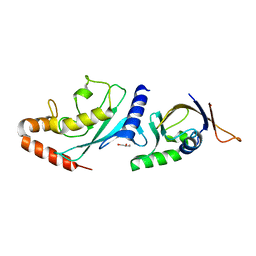 | | Structure of the A.thaliana E1 UFD domain in complex with E2 | | Descriptor: | GLYCEROL, SAE2, SUMO-conjugating enzyme SCE1 | | Authors: | Liu, B, Lois, L.M, Reverter, D. | | Deposit date: | 2018-06-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into SUMO E1-E2 interactions in Arabidopsis uncovers a distinctive platform for securing SUMO conjugation specificity across evolution.
Biochem.J., 476, 2019
|
|
6GV3
 
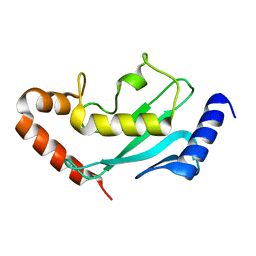 | |
3EH4
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
3EH3
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
3EH5
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
3QJV
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJR
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJS
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJQ
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJU
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJT
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
5IPN
 
 | | SigmaS-transcription initiation complex with 4-nt nascent RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-03-30 | | Last modified: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (4.61 Å) | | Cite: | Structures of E. coli sigma S-transcription initiation complexes provide new insights into polymerase mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IPL
 
 | | SigmaS-transcription initiation complex with 4-nt nascent RNA | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of E. coli sigma S-transcription initiation complexes provide new insights into polymerase mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IPM
 
 | | SigmaS-transcription initiation complex with 4-nt nascent RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-03-30 | | Last modified: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structures of E. coli sigma S-transcription initiation complexes provide new insights into polymerase mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2BB4
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Asp-Phe at pH 5.0 | | Descriptor: | ASPARTIC ACID, CALCIUM ION, Elastase-1, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-17 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BDB
 
 | | Porcine pancreatic elastase complexed with Asn-Pro-Ile and Ala-Ala at pH 5.0 | | Descriptor: | ALANINE, CALCIUM ION, Elastase-1, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BDA
 
 | | Porcine pancreatic elastase complexed with N-acetyl-NPI and Ala-Ala at pH 5.0 | | Descriptor: | ALANINE, CALCIUM ION, Elastase-1, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BDC
 
 | |
