1O91
 
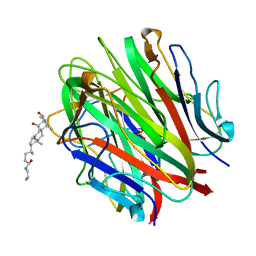 | | Crystal Structure of a Collagen VIII NC1 Domain Trimer | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, COLLAGEN ALPHA 1(VIII) CHAIN, SULFATE ION | | Authors: | Kvansakul, M, Bogin, O, Hohenester, E, Yayon, A. | | Deposit date: | 2002-12-10 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Collagen Alpha1(Viii) Nc1 Trimer.
Matrix Biol., 22, 2003
|
|
5KK4
 
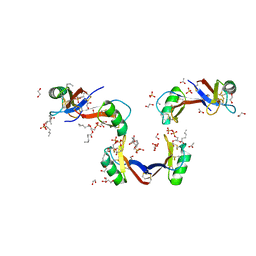 | | Crystal Structure of the Plant Defensin NsD7 bound to Phosphatidic Acid | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kvansakul, M, Hulett, M.D, Lay, F.T. | | Deposit date: | 2016-06-21 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of phosphatidic acid by NsD7 mediates the formation of helical defensin-lipid oligomeric assemblies and membrane permeabilization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1GL4
 
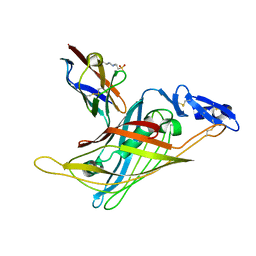 | | Nidogen-1 G2/Perlecan IG3 Complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BASEMENT MEMBRANE-SPECIFIC HEPARAN SULFATE PROTEOGLYCAN CORE PROTEIN, NIDOGEN-1, ... | | Authors: | Kvansakul, M, Hopf, M, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the High-Affinity Interaction of Nidogen-1 with Immunoglobulin-Like Domain 3 of Perlecan
Embo J., 20, 2001
|
|
1UX6
 
 | |
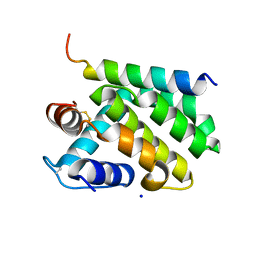
2V6Q
 
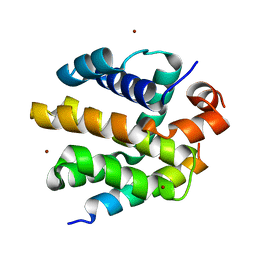 | |
2VTY
 
 | | Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain swapped dimer | | Descriptor: | PROTEIN F1 | | Authors: | Kvansakul, M, Yang, H, Fairlie, W.D, Czabotar, P.E, Fischer, S.F, Perugini, M.A, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2008-05-19 | | Release date: | 2008-06-24 | | Last modified: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vaccinia Virus Anti-Apoptotic F1L is a Novel Bcl-2-Like Domain-Swapped Dimer that Binds a Highly Selective Subset of Bh3-Containing Death Ligands.
Cell Death Differ., 15, 2008
|
|
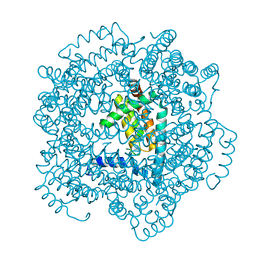
2JBX
 
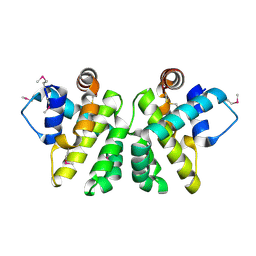 | | Crystal Structure of the myxoma virus anti-apoptotic protein M11L | | Descriptor: | M11L PROTEIN | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Structural Viral Mimic of Prosurvival Bcl-2: A Pivotal Role for Sequestering Proapoptotic Bax and Bak.
Mol.Cell, 25, 2007
|
|
2JBY
 
 | | A viral protein unexpectedly mimics the structure and function of pro- survival Bcl-2 | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER 2, M11L PROTEIN, SODIUM ION | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak.
Mol. Cell, 25, 2007
|
|
2WH6
 
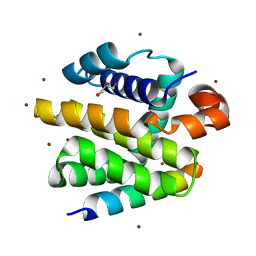 | | Crystal structure of anti-apoptotic BHRF1 in complex with the Bim BH3 domain | | Descriptor: | BCL-2-LIKE PROTEIN 11, BROMIDE ION, EARLY ANTIGEN PROTEIN R, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2009-05-01 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1.
PLoS Pathog., 6, 2010
|
|
2XPX
 
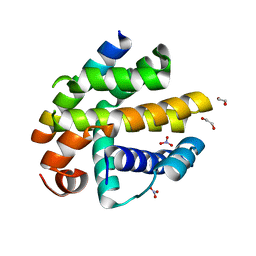 | | Crystal structure of BHRF1:Bak BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BHRF1, BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1.
Plos Pathog., 6, 2010
|
|
7T9Q
 
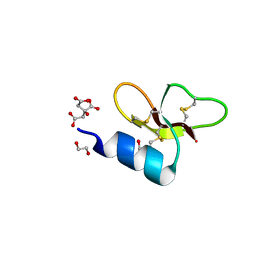 | | Crystal structure of Crocodile defensin CpoBD13 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, CpoBD13 | | Authors: | Kvansakul, M, Williams, S, Hulett, M.D. | | Deposit date: | 2021-12-19 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crocodile defensin (CpoBD13) antifungal activity via pH-dependent phospholipid targeting and membrane disruption.
Nat Commun, 14, 2023
|
|
7T9R
 
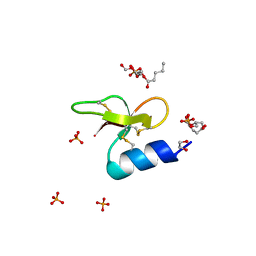 | | Crystal structure of Crocodile defensin CpoBD13:phosphatidic acid complex | | Descriptor: | (R)-2-(FORMYLOXY)-3-(PHOSPHONOOXY)PROPYL PENTANOATE, 1,2-ETHANEDIOL, CpoBD13, ... | | Authors: | Kvansakul, M, Williams, S, Hulett, M.D. | | Deposit date: | 2021-12-19 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crocodile defensin (CpoBD13) antifungal activity via pH-dependent phospholipid targeting and membrane disruption.
Nat Commun, 14, 2023
|
|
6WH0
 
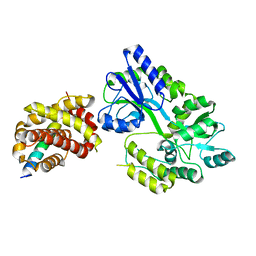 | | Crystal structure of HyBcl-2-4 with HyBax BH3 | | Descriptor: | Apoptosis regulator BAX, Maltodextrin-binding protein,Bcl-2-like 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kvansakul, M, Hinds, M.G, Banjara, S. | | Deposit date: | 2020-04-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structural basis of Bcl-2 mediated cell death regulation in hydra.
Biochem.J., 477, 2020
|
|
6WGZ
 
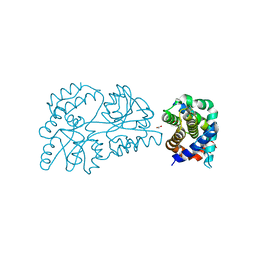 | |
5AEA
 
 | |
5AJJ
 
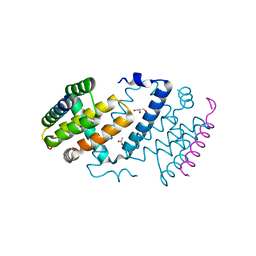 | |
5AJK
 
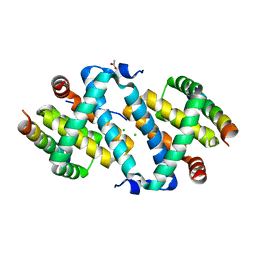 | |
4D2M
 
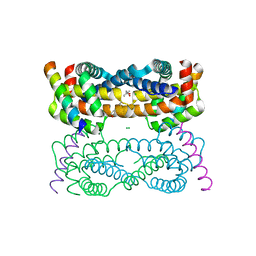 | | Vaccinia Virus F1L bound to Bim BH3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, BCL-2-LIKE PROTEIN 11, CHLORIDE ION, ... | | Authors: | Kvansakul, M, Colman, P.M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insight Into Bh3-Domain Binding of Vaccinia Virus Anti-Apoptotic F1L.
J.Virol., 88, 2014
|
|
4D2L
 
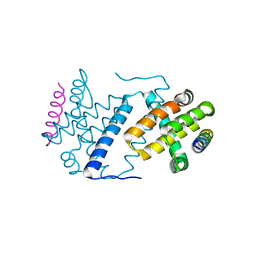 | | Vaccinia Virus F1L bound to Bak BH3 | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, PROTEIN F1L | | Authors: | Kvansakul, M, Colman, P.M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural Insight Into Bh3-Domain Binding of Vaccinia Virus Anti-Apoptotic F1L.
J.Virol., 88, 2014
|
|
7JO7
 
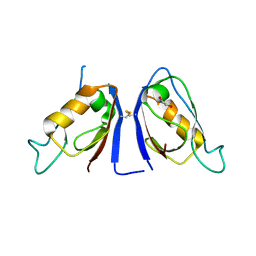 | | Crystal Structure of Human Scribble PDZ2 | | Descriptor: | 1,2-ETHANEDIOL, Protein scribble homolog | | Authors: | How, J.Y, Kvansakul, M. | | Deposit date: | 2020-08-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis of the human Scribble-Vangl2 association in health and disease.
Biochem.J., 478, 2021
|
|
6B55
 
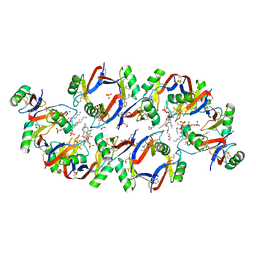 | | Crystal structure of the Plant Defensin NaD1 complexed with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, 1,2-ETHANEDIOL, Flower-specific defensin, ... | | Authors: | Jarva, M, Phan, K, Humble, C, Lay, F.T, Hulett, M, Kvansakul, M. | | Deposit date: | 2017-09-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of a carpet-like antimicrobial defensin-phospholipid membrane disruption complex.
Nat Commun, 9, 2018
|
|
6CS9
 
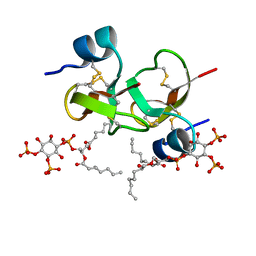 | | Crystal structure of human beta-defensin 2 in complex with PIP2 | | Descriptor: | Beta-defensin 4A, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Jarva, M, Phan, K, Lay, F.T, Humble, C, Hulett, M, Kvansakul, M. | | Deposit date: | 2018-03-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human beta-defensin 2 killsCandida albicansthrough phosphatidylinositol 4,5-bisphosphate-mediated membrane permeabilization.
Sci Adv, 4, 2018
|
|
4UF1
 
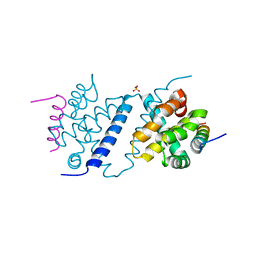 | | Deerpox virus DPV022 in complex with Bak BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UF2
 
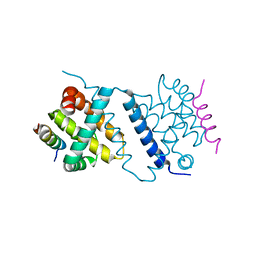 | | Deerpox virus DPV022 in complex with Bax BH3 | | Descriptor: | Antiapoptotic membrane protein, Apoptosis regulator BAX | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UF3
 
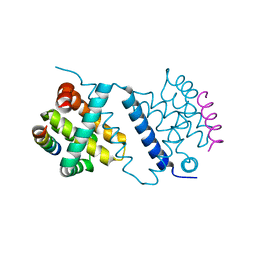 | | Deerpox virus DPV022 in complex with Bim BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2-like protein 11 | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
