8DWN
 
 | |
3RGK
 
 | | Crystal Structure of Human Myoglobin Mutant K45R | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hubbard, S.R. | | Deposit date: | 2011-04-08 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of a recombinant human myoglobin mutant at 2.8 A resolution
J.Mol.Biol., 20, 1990
|
|
1IR3
 
 | |
1IRK
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HUMAN INSULIN RECEPTOR | | Descriptor: | ETHYL MERCURY ION, INSULIN RECEPTOR TYROSINE KINASE DOMAIN | | Authors: | Hubbard, S.R, Wei, L, Ellis, L, Hendrickson, W.A. | | Deposit date: | 1995-01-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of the human insulin receptor.
Nature, 372, 1994
|
|
7TEU
 
 | | Crystal structure of JAK2 JH1 with type II inhibitor YLIU-04-105-1 | | Descriptor: | 3-{(4S)-2-[(cyclopropanecarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}-4-methylbenzamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Hubbard, S.R. | | Deposit date: | 2022-01-05 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New scaffolds for type II JAK2 inhibitors overcome the acquired G993A resistance mutation.
Cell Chem Biol, 30, 2023
|
|
6D42
 
 | |
3HK0
 
 | |
4XLV
 
 | |
8EX1
 
 | |
8EX0
 
 | |
8EX2
 
 | |
1AGW
 
 | |
4K81
 
 | |
3ML4
 
 | |
3HKL
 
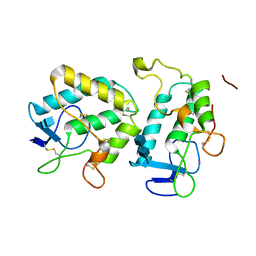 | | Crystal Structure of the Frizzled-like Cysteine-rich Domain of MuSK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Muscle, skeletal receptor tyrosine protein kinase | | Authors: | Stiegler, A.L, Hubbard, S.R. | | Deposit date: | 2009-05-24 | | Release date: | 2009-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the frizzled-like cysteine-rich domain of the receptor tyrosine kinase MuSK.
J.Mol.Biol., 393, 2009
|
|
3TCA
 
 | |
1YVH
 
 | |
3BCH
 
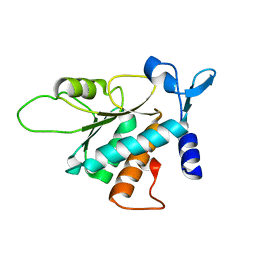 | |
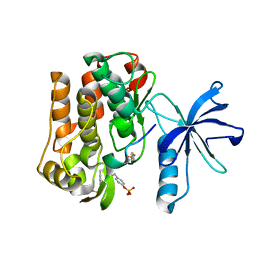
3BU3
 
 | |
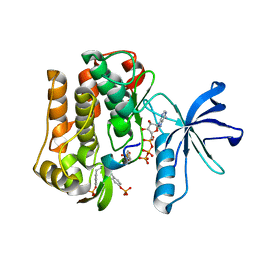
3BU5
 
 | |
2B4S
 
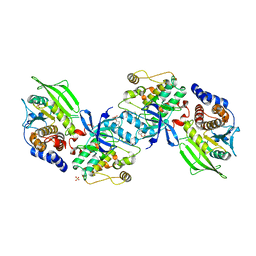 | | Crystal structure of a complex between PTP1B and the insulin receptor tyrosine kinase | | Descriptor: | Insulin receptor, SULFATE ION, Tyrosine-protein phosphatase, ... | | Authors: | Li, S, Depetris, R.S, Barford, D, Chernoff, J, Hubbard, S.R. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Complex between Protein Tyrosine Phosphatase 1B and the Insulin Receptor Tyrosine Kinase.
Structure, 13, 2005
|
|
2AUG
 
 | | Crystal structure of the Grb14 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 14 | | Authors: | Depetris, R.S, Hu, J, Gimpelevich, I, Holt, L.J, Daly, R.J, Hubbard, S.R. | | Deposit date: | 2005-08-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the insulin receptor by the adaptor protein grb14.
Mol.Cell, 20, 2005
|
|
3BU6
 
 | |
2AUH
 
 | | Crystal structure of the Grb14 BPS region in complex with the insulin receptor tyrosine kinase | | Descriptor: | CALCIUM ION, Growth factor receptor-bound protein 14, Insulin receptor | | Authors: | Depetris, R.S, Hu, J, Gimpelevich, I, Holt, L.J, Daly, R.J, Hubbard, S.R. | | Deposit date: | 2005-08-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of the insulin receptor by the adaptor protein grb14.
Mol.Cell, 20, 2005
|
|
3D94
 
 | | Crystal structure of the insulin-like growth factor-1 receptor kinase in complex with PQIP | | Descriptor: | 3-[cis-3-(4-methylpiperazin-1-yl)cyclobutyl]-1-(2-phenylquinolin-7-yl)imidazo[1,5-a]pyrazin-8-amine, CALCIUM ION, Insulin-like growth factor 1 receptor beta chain | | Authors: | Wu, J, Li, W, Miller, W.T, Hubbard, S.R. | | Deposit date: | 2008-05-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor
Embo J., 27, 2008
|
|
