4S05
 
 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (26-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
4S04
 
 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (25-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
1E9L
 
 | |
1C72
 
 | | TYR115, GLN165 AND TRP209 CONTRIBUTE TO THE 1,2-EPOXY-3-(P-NITROPHENOXY)PROPANE CONJUGATING ACTIVITIES OF GLUTATHIONE S-TRANSFERASE CGSTM1-1 | | Descriptor: | 1-HYDROXY-2-S-GLUTATHIONYL-3-PARA-NITROPHENOXY-PROPANE, PROTEIN (GLUTATHIONE S-TRANSFERASE) | | Authors: | Chern, M.K, Wu, T.C, Hsieh, C.H, Chou, C.C, Liu, L.F, Kuan, I.C, Yeh, Y.H, Hsiao, C.D, Tam, M.F. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tyr115, gln165 and trp209 contribute to the 1, 2-epoxy-3-(p-nitrophenoxy)propane-conjugating activity of glutathione S-transferase cGSTM1-1.
J.Mol.Biol., 300, 2000
|
|
7DV3
 
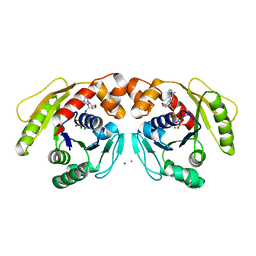 | | Structure of Sulfolobus solfataricus SegA-AMPPNP protein | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SOJ protein (Soj) | | Authors: | Yen, C.Y, Lin, M.G, Wu, C.T, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DUV
 
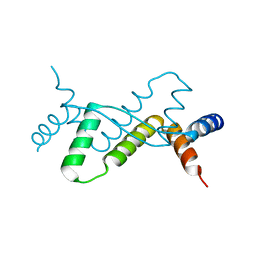 | | Structure of Sulfolobus solfataricus SegB protein | | Descriptor: | SULFATE ION, SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DUT
 
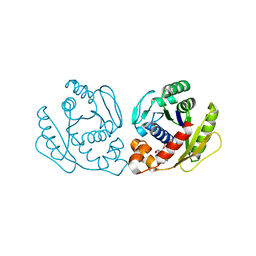 | | Structure of Sulfolobus solfataricus SegA protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SOJ protein (Soj) | | Authors: | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV2
 
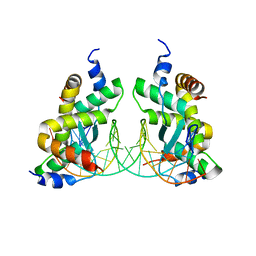 | | Structure of Sulfolobus solfataricus SegB-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*AP*GP*AP*AP*GP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*CP*TP*TP*CP*TP*AP*CP*GP*TP*A)-3'), SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DWR
 
 | | Structure of Sulfolobus solfataricus SegA-ADP complex bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
2W58
 
 | | Crystal Structure of the DnaI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, K.L, Lo, Y.H, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Interplay between the Replicative Helicase Dnac and its Loader Protein Dnai from Geobacillus Kaustophilus.
J.Mol.Biol., 393, 2009
|
|
6JSX
 
 | |
4AWX
 
 | | Moonlighting functions of FeoC in the regulation of ferrous iron transport in Feo | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, FERROUS IRON TRANSPORT PROTEIN C, NICKEL (II) ION, ... | | Authors: | Hung, K.-W, Tsai, J.-Y, Juan, T.-H, Hsu, Y.-L, Hsiao, C.D, Huang, T.-H. | | Deposit date: | 2012-06-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Klebsiella Pneumoniae Nfeob/Feoc Complex and Roles of Feoc in Regulation of Fe2+ Transport by the Bacterial Feo System.
J.Bacteriol., 194, 2012
|
|
4NHJ
 
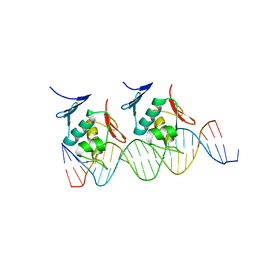 | | Crystal structure of Klebsiella pneumoniae RstA DNA-binding domain in complex with RstA box | | Descriptor: | 5'-D(*CP*AP*GP*GP*GP*AP*GP*TP*AP*AP*CP*GP*GP*AP*AP*TP*GP*TP*AP*CP*AP*AP*C)-3', 5'-D(*GP*GP*TP*TP*GP*TP*AP*CP*AP*TP*TP*CP*CP*GP*TP*TP*AP*CP*TP*CP*CP*CP*T)-3', DNA-binding transcriptional regulator RstA | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2013-11-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
4NIC
 
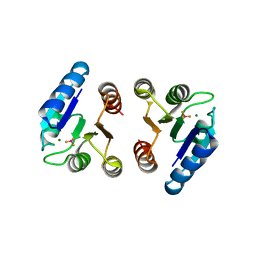 | |
4OEG
 
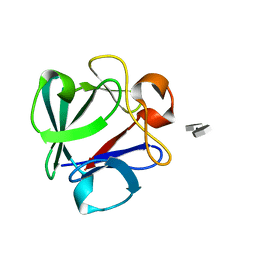 | | Crystal Structure Analysis of FGF2-Disaccharide (S9I2) complex | | Descriptor: | 2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
4OEF
 
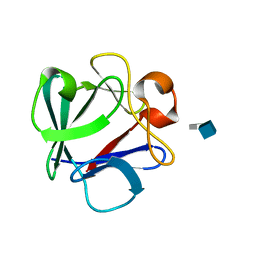 | | Crystal Structure Analysis of FGF2-Disaccharide (S6I2) complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
4OEE
 
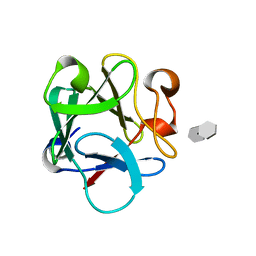 | | Crystal Structure Analysis of FGF2-Disaccharide (S3I2) complex | | Descriptor: | 2-deoxy-3-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
1H65
 
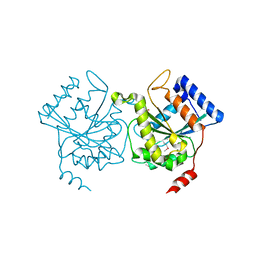 | | Crystal structure of pea Toc34 - a novel GTPase of the chloroplast protein translocon | | Descriptor: | CHLOROPLAST OUTER ENVELOPE PROTEIN OEP34, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, Y.J, Forouhar, F, Li, H.M, Tu, S.L, Kao, S, Shr, H.L, Chou, C.C, Hsiao, C.D. | | Deposit date: | 2001-06-06 | | Release date: | 2002-01-29 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pea Toc34 - a Novel Gtpase of the Chloroplast Protein Translocon
Nat.Struct.Biol., 9, 2002
|
|
5WTN
 
 | |
2VYE
 
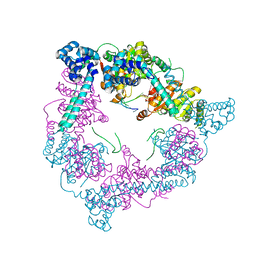 | | Crystal Structure of the DnaC-ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP)-3', REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
2VYF
 
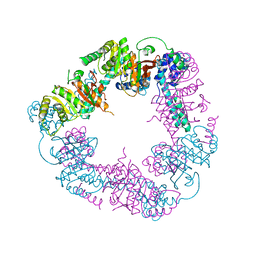 | | Crystal Structure of the DnaC | | Descriptor: | GOLD ION, REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
5ZME
 
 | |
5ZMF
 
 | | AMPPNP complex of C. reinhardtii ArsA1 | | Descriptor: | ATPase ARSA1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lin, T.W, Hsiao, C.D, Chang, H.Y. | | Deposit date: | 2018-04-03 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.556 Å) | | Cite: | Structural analysis of chloroplast tail-anchored membrane protein recognition by ArsA1.
Plant J., 99, 2019
|
|
1UD0
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL 10-kDA SUBDOMAIN OF HSC70 | | Descriptor: | 70 kDa heat-shock-like protein, SODIUM ION | | Authors: | Chou, C.C, Forouhar, F, Yeh, Y.H, Wang, C, Hsiao, C.D. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the C-terminal 10-kDa subdomain of Hsc70
J.BIOL.CHEM., 278, 2003
|
|
1GGG
 
 | |
