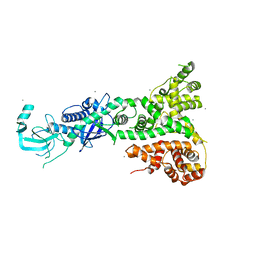
2FFL
 
 | |
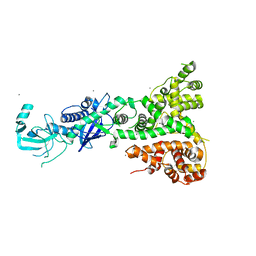
2QVW
 
 | |
8VXA
 
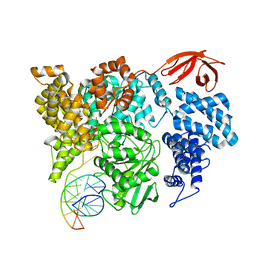 | |
8VX9
 
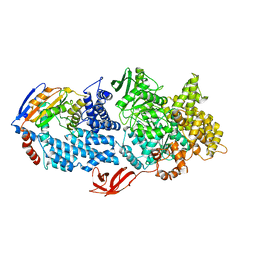 | |
8VXC
 
 | |
8VXY
 
 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | Authors: | Tuck, O.T, Hu, J.J, Doudna, J.A. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
6VPC
 
 | | Structure of the SpCas9 DNA adenine base editor - ABE8e | | Descriptor: | CRISPR-associated endonuclease Cas9, Cas9 (SpCas9) single-guide RNA (sgRNA), DNA non-target strand (NTS), ... | | Authors: | Knott, G.J, Lapinaite, A, Doudna, J.A. | | Deposit date: | 2020-02-03 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | DNA capture by a CRISPR-Cas9-guided adenine base editor.
Science, 369, 2020
|
|
7KFU
 
 | | Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
7KFT
 
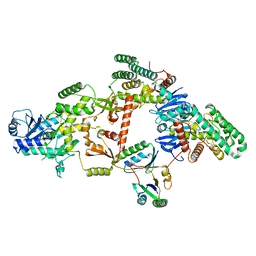 | | Partial Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
7S38
 
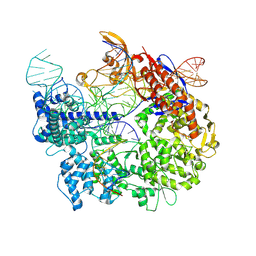 | | Cas9:sgRNA:DNA (S. pyogenes) forming a 3-base-pair R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S36
 
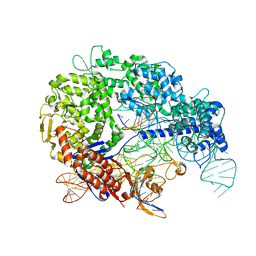 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, closed-protein/bent-DNA conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S37
 
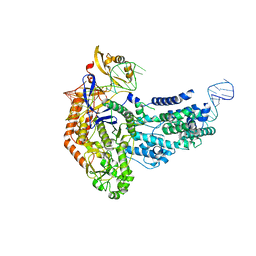 | | Cas9:sgRNA (S. pyogenes) in the open-protein conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Single-guide RNA | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S3H
 
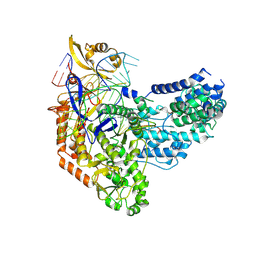 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, open-protein/linear-DNA conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-06 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7THB
 
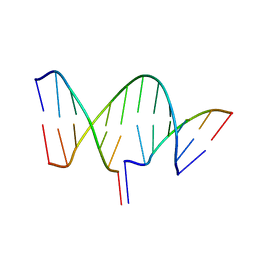 | | Crystal structure of an RNA-5'/DNA-3' strand exchange junction | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*TP*AP*AP*GP*CP*AP*GP*CP*AP*TP*C)-3'), RNA (5'-R(*AP*GP*CP*UP*UP*AP*C)-3') | | Authors: | Cofsky, J.C, Knott, G.J, Gee, C.L, Doudna, J.A. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of an RNA/DNA strand exchange junction.
Plos One, 17, 2022
|
|
4TXY
 
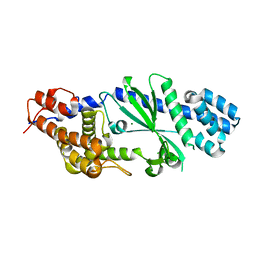 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase, a prokaryotic cGAS homolog | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0001 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
4TY0
 
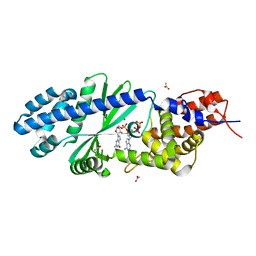 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with linear intermediate 5' pppA(3',5')pG | | Descriptor: | ACETATE ION, Cyclic AMP-GMP synthase, MAGNESIUM ION, ... | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
4TXZ
 
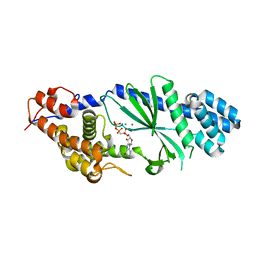 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with nonhydrolyzable GTP | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
1GID
 
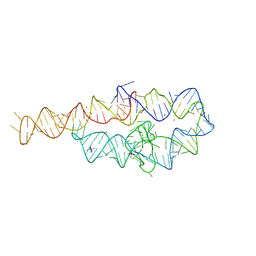 | | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, P4-P6 RNA RIBOZYME DOMAIN | | Authors: | Cate, J.H, Gooding, A.R, Podell, E, Zhou, K, Golden, B.L, Kundrot, C.E, Cech, T.R, Doudna, J.A. | | Deposit date: | 1996-08-22 | | Release date: | 1996-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a group I ribozyme domain: principles of RNA packing.
Science, 273, 1996
|
|
4WYQ
 
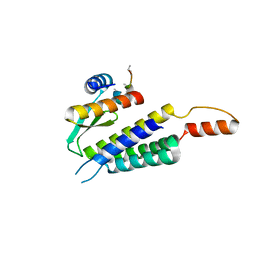 | | Crystal structure of the Dicer-TRBP interface | | Descriptor: | Endoribonuclease Dicer, Poly(UNK), RISC-loading complex subunit TARBP2 | | Authors: | Wilson, R.C, Doudna, J.A. | | Deposit date: | 2014-11-18 | | Release date: | 2014-12-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis.
Mol.Cell, 57, 2015
|
|
4KM5
 
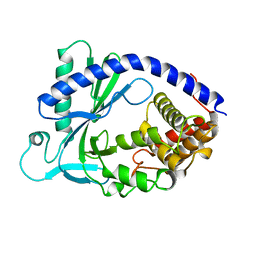 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
3FRX
 
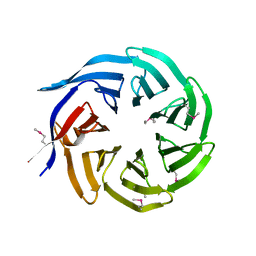 | |
2OIH
 
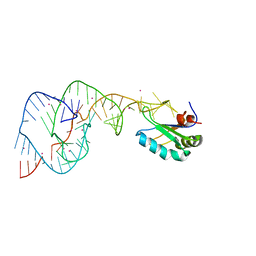 | | Hepatitis Delta Virus gemonic ribozyme precursor with C75U mutation and bound to monovalent cation Tl+ | | Descriptor: | HDV ribozyme, THALLIUM (I) ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Ding, F, Batchelor, J.D, Doudna, J.A. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural roles of monovalent cations in the HDV ribozyme.
Structure, 15, 2007
|
|
2OJ3
 
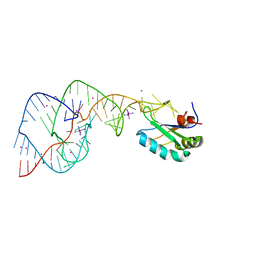 | | Hepatitis Delta Virus ribozyme precursor structure, with C75U mutation, bound to Tl+ and cobalt hexammine (Co(NH3)63+) | | Descriptor: | COBALT HEXAMMINE(III), HDV RIBOZYME, THALLIUM (I) ION, ... | | Authors: | Ke, A, Ding, F, Batchelor, J.D, Doudna, J.A. | | Deposit date: | 2007-01-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural roles of monovalent cations in the HDV ribozyme.
Structure, 15, 2007
|
|
6MCC
 
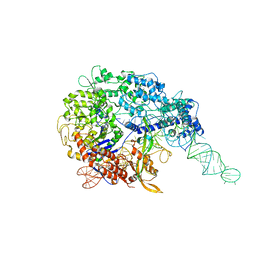 | |
6MCB
 
 | |
