8JTP
 
 | |
7CK4
 
 | |
3O85
 
 | | Giardia lamblia 15.5kD RNA binding protein | | Descriptor: | Ribosomal protein L7Ae | | Authors: | Biswas, S, Maxwell, E.S. | | Deposit date: | 2010-08-02 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Comparative analysis of the 15.5kD box C/D snoRNP core protein in the primitive eukaryote Giardia lamblia reveals unique structural and functional features.
Biochemistry, 50, 2011
|
|
3PAF
 
 | | M. jannaschii L7Ae mutant | | Descriptor: | 50S ribosomal protein L7Ae, ACETATE ION, SULFATE ION | | Authors: | Biswas, S, Maxwell, E.S. | | Deposit date: | 2010-10-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability of M.jannaschii L7Ae El9 KtoQ mutant
To be Published
|
|
3R17
 
 | |
3R16
 
 | | Human CAII bound to N-(4-sulfamoylphenyl)-2-(thiophen-2-yl) acetamide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biswas, S, McKenna, R, Supuran, C.T. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational variability of different sulfonamide inhibitors with thienyl-acetamido moieties attributes to differential binding in the active site of cytosolic human carbonic anhydrase isoforms.
Bioorg.Med.Chem., 19, 2011
|
|
2ODJ
 
 | |
2QTK
 
 | |
4ILX
 
 | |
4IWZ
 
 | | structure of hCAII in complex with an acetazolamide derivative | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biswas, S, McKenna, R. | | Deposit date: | 2013-01-24 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Effect of incorporating a thiophene tail in the scaffold of acetazolamide on the inhibition of human carbonic anhydrase isoforms I, II, IX and XII.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4K0Z
 
 | |
4K13
 
 | |
4K1Q
 
 | |
4K0T
 
 | |
4K0S
 
 | |
1IWD
 
 | |
5Z5O
 
 | | Structure of Pycnonodysostosis disease related I249T mutant of human cathepsin K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Biswas, S, Roy, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Not all pycnodysostosis-related mutants of human cathepsin K are inactive - crystal structure and biochemical studies of an active mutant I249T.
FEBS J., 285, 2018
|
|
3T7Z
 
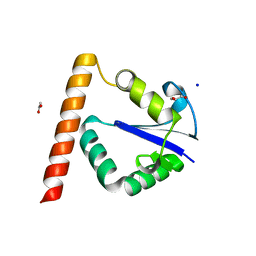 | | Structure of Methanocaldococcus jannaschii Nop N-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleolar protein Nop 56/58, ... | | Authors: | Biswas, S, Maxwell, S. | | Deposit date: | 2011-07-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structurally Conserved Nop56/58 N-terminal Domain Facilitates Archaeal Box C/D Ribonucleoprotein-guided Methyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
4ITP
 
 | | Structure of human carbonic anhydrase II bound to a benzene sulfonamide | | Descriptor: | 2-phenyl-N-(4-sulfamoylbenzyl)acetamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Biswas, S, McKenna, R. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Structural study of the location of the phenyl tail of benzene sulfonamides and the effect on human carbonic anhydrase inhibition.
Bioorg.Med.Chem., 21, 2013
|
|
7KNL
 
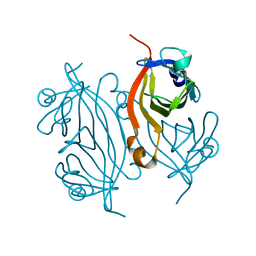 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | Streptavidin | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Biswas, A, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-11-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
7KBY
 
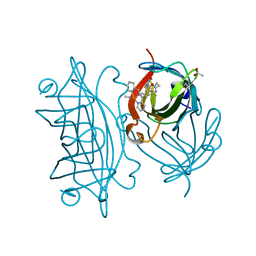 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, CYANIDE ION, Streptavidin, ... | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
7KBZ
 
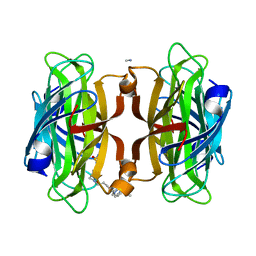 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | CYANIDE ION, Streptavidin, {N-(4-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}butyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
4FJR
 
 | | Mode of interaction of Merocyanine 540 with HEW Lysozyme | | Descriptor: | 3-[(2E)-2-[(2Z)-4-(1,3-dibutyl-4,6-dioxo-2-thioxotetrahydropyrimidin-5(2H)-ylidene)but-2-en-1-ylidene]-1,3-benzoxazol-3(2H)-yl]propane-1-sulfonic acid, CHLORIDE ION, Lysozyme C | | Authors: | Mitra, P, Banerjee, M, Basu, S, Biswas, S. | | Deposit date: | 2012-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mode of interaction of Merocyanine 540 dye with lysozyme
To be Published
|
|
6XLQ
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
8GT7
 
 | | Structure of falcipain and human Stefin A mutant complex | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Cystatin-A, ... | | Authors: | Chakraborty, S, Biswas, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structure of falcipain and human Stefin A complex
To Be Published
|
|
