3W8I
 
 | |
3W8H
 
 | |
8AN4
 
 | | MenT1 toxin (rv0078a) from Mycobacterium tuberculosis H37Rv | | Descriptor: | Bacterial toxin | | Authors: | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
8AN5
 
 | | MenAT1 toxin-antitoxin complex (rv0078a-rv0078b) from Mycobacterium tuberculosis H37Rv | | Descriptor: | Bacterial toxin, Conserved protein | | Authors: | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
3VCB
 
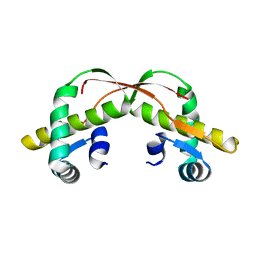 | | C425S mutant of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3VC8
 
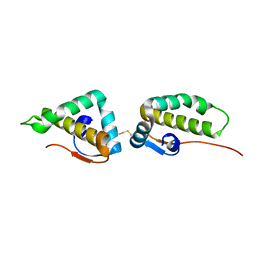 | | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3BEQ
 
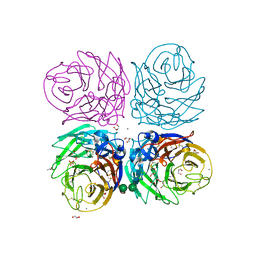 | | Neuraminidase of A/Brevig Mission/1/1918 H1N1 strain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, X, Zhu, X, Wilson, I.A. | | Deposit date: | 2007-11-19 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of the 1918 influenza virus H1N1 neuraminidase
J.Virol., 82, 2008
|
|
3B7E
 
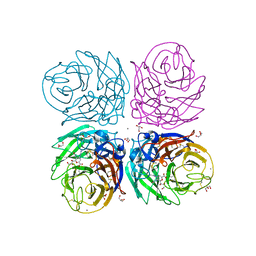 | | Neuraminidase of A/Brevig Mission/1/1918 H1N1 strain in complex with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Xu, X, Zhu, X, Wilson, I.A. | | Deposit date: | 2007-10-30 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural characterization of the 1918 influenza virus H1N1 neuraminidase
J.Virol., 82, 2008
|
|
1T09
 
 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex NADP | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
1T0L
 
 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex with NADP, isocitrate, and calcium(2+) | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-10 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
7S0R
 
 | |
4OIV
 
 | | Structural basis for small molecule NDB as a selective antagonist of FXR | | Descriptor: | Bile acid receptor, N-benzyl-N-(3-tert-butyl-4-hydroxyphenyl)-2,6-dichloro-4-(dimethylamino)benzamide | | Authors: | Xu, X, Chen, L, Hu, L, Shen, X. | | Deposit date: | 2014-01-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Small Molecule NDB (N-Benzyl-N-(3-(tert-butyl)-4-hydroxyphenyl)-2,6-dichloro-4-(dimethylamino) Benzamide) as a Selective Antagonist of Farnesoid X Receptor alpha (FXR alpha ) in Stabilizing the Homodimerization of the Receptor.
J.Biol.Chem., 290, 2015
|
|
2GTH
 
 | | crystal structure of the wildtype MHV coronavirus non-structural protein nsp15 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
2GTI
 
 | | mutation of MHV coronavirus non-structural protein nsp15 (F307L) | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, SULFATE ION | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
4ZLP
 
 | | Crystal Structure of Notch3 Negative Regulatory Region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2015-05-01 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Insights into Autoregulation of Notch3 from Structural and Functional Studies of Its Negative Regulatory Region.
Structure, 23, 2015
|
|
4ZSO
 
 | | Crystal structure of a complex between B7-H6, a tumor cell ligand for natural cytotoxicity receptor NKp30, and an inhibitory antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, X, Li, Y, Mariuzza, R.A. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a complex between B7-H6, a tumor cell ligand for natural cytotoxicity receptor NKp30, and an inhibitory antibody
to be published
|
|
1PCW
 
 | | Aquifex aeolicus KDO8PS in complex with cadmium and APP, a bisubstrate inhibitor | | Descriptor: | 1-DEOXY-6-O-PHOSPHONO-1-[(PHOSPHONOMETHYL)AMINO]-L-THREO-HEXITOL, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION | | Authors: | Xu, X, Wang, J, Grison, C, Petek, S, Coutrot, P, Birck, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2003-05-17 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of Novel Inhibitors of 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase.
Drug DES.DISCOVERY, 18, 2003
|
|
1ZHA
 
 | | A. aeolicus KDO8PS R106G mutant in complex with PEP and R5P | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop.
Biochemistry, 44, 2005
|
|
1ZJI
 
 | | Aquifex aeolicus KDO8PS R106G mutant in complex with 2PGA and R5P | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop
Biochemistry, 44, 2005
|
|
6UWR
 
 | |
6UWT
 
 | |
1TER
 
 | |
8XOO
 
 | |
8XOP
 
 | |
8XON
 
 | |
