1AMO
 
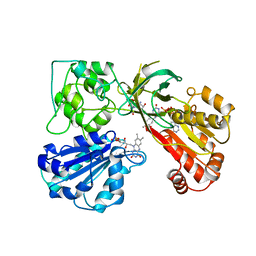 | | THREE-DIMENSIONAL STRUCTURE OF NADPH-CYTOCHROME P450 REDUCTASE: PROTOTYPE FOR FMN-AND FAD-CONTAINING ENZYMES | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wang, M, Roberts, D.L, Paschke, R, Shea, T.M, Masters, B.S.S, Kim, J.J.P. | | Deposit date: | 1997-06-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
4QY0
 
 | | Structure of H10 from human-infecting H10N8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY2
 
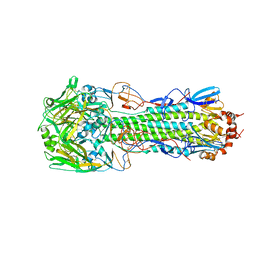 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4X1D
 
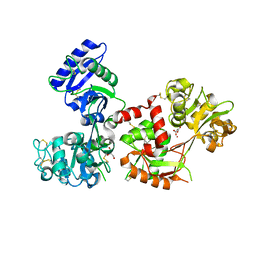 | | Ytterbium-bound human serum transferrin | | Descriptor: | GLYCEROL, MALONATE ION, Serotransferrin, ... | | Authors: | Wang, M, Zhang, H, Sun, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | "Anion clamp" allows flexible protein to impose coordination geometry on metal ions
Chem.Commun.(Camb.), 51, 2015
|
|
4X1B
 
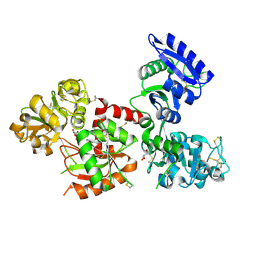 | | Human serum transferrin with ferric ion bound at the C-lobe only | | Descriptor: | FE (III) ION, GLYCEROL, MALONATE ION, ... | | Authors: | Wang, M, Zhang, H, Sun, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | "Anion clamp" allows flexible protein to impose coordination geometry on metal ions
Chem.Commun.(Camb.), 51, 2015
|
|
3NCI
 
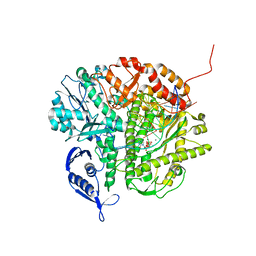 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite dG at 1.8 angstrom resolution | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Blaha, G, Steitz, T.A, Konigsberg, W.H, Wang, J. | | Deposit date: | 2010-06-04 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Insights into base selectivity from the 1.8 A resolution structure of an RB69 DNA polymerase ternary complex.
Biochemistry, 50, 2011
|
|
8GPS
 
 | |
8GPV
 
 | |
6JJP
 
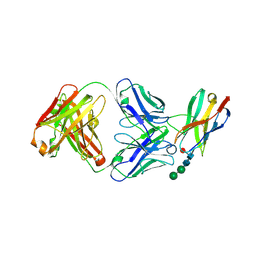 | | Crystal structure of Fab of a PD-1 monoclonal antibody MW11-h317 in complex with PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of MW11-h317, Programmed cell death protein 1, ... | | Authors: | Wang, M, Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation.
Commun Biol, 2, 2019
|
|
1NHV
 
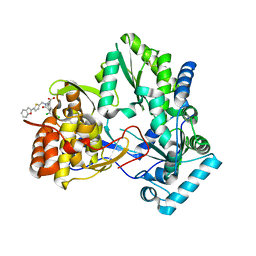 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(5-BENZOFURAN-2-YL-THIOPHEN-2-YLMETHYL)-(2,4-DICHLORO-BENZOYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
1NHU
 
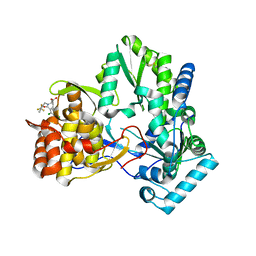 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
7FFM
 
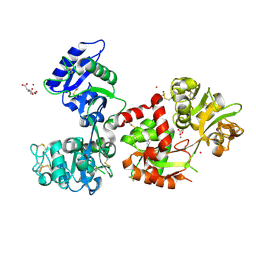 | | Human serum transferrin with five osmium binding sites | | Descriptor: | MALONATE ION, NITRILOTRIACETIC ACID, OSMIUM ION, ... | | Authors: | Wang, M, Sun, H. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
7FFU
 
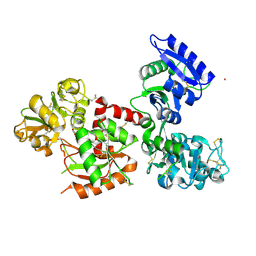 | | Osmium-bound human serum transferrin | | Descriptor: | FE (III) ION, MALONATE ION, OSMIUM ION, ... | | Authors: | Wang, M, Sun, H. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
2FXZ
 
 | |
2FXY
 
 | |
2KQ3
 
 | | Solution structure of SNase140 | | Descriptor: | Thermonuclease | | Authors: | Wang, M, Feng, Y, Yao, H, Wang, J. | | Deposit date: | 2009-10-26 | | Release date: | 2010-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Importance of the C-Terminal Loop L137-S141 for the Folding and Folding Stability of Staphylococcal Nuclease
Biochemistry, 49, 2010
|
|
2KJ4
 
 | | Solution structure of the complex of VEK-30 and plasminogen kringle 2 | | Descriptor: | VEK-30, plasminogen | | Authors: | Wang, M, Zajicek, J, Prorok, M, Castellin, F.J. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex of VEK-30 and plasminogen kringle 2.
J.Struct.Biol., 169, 2010
|
|
7VGA
 
 | | Cryo-EM structure of alphavirus, Getah virus | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Wang, M, Sun, Z.Z, Wang, J.F. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Implications for the pathogenicity and antigenicity of alpha viruses revealed by a 3.5 angstrom Cryo-EM structure of Getah virus
To Be Published
|
|
7WCO
 
 | | Cryo-EM structure of alphavirus, Getah virus | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Wang, M, Sun, Z.Z, Wang, J.F. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Implications for the pathogenicity and antigenicity of alpha viruses revealed by a 3.5 angstrom Cryo-EM structure of Getah virus
To Be Published
|
|
7WC2
 
 | | Cryo-EM structure of alphavirus, Getah virus | | Descriptor: | Spike glycoprotein E1, Spike glycoprotein E2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, M, Sun, Z.Z, Wang, J.F. | | Deposit date: | 2021-12-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implications for the pathogenicity and antigenicity of alpha viruses revealed by a 3.5 angstrom Cryo-EM structure of Getah virus
To Be Published
|
|
5ZQY
 
 | | Crystal structure of a poly(ADP-ribose) glycohydrolase | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, M, Yuan, Z, Ma, Y, Wang, J, Liu, X. | | Deposit date: | 2018-04-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Structure-function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation.
J. Biol. Chem., 293, 2018
|
|
6JAS
 
 | | Human serum transferrin with iron citrate bound | | Descriptor: | CITRIC ACID, FE (III) ION, MALONATE ION, ... | | Authors: | Wang, M, Wang, H, Sun, H. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Binding of ruthenium and osmium at non-iron sites of transferrin explains their iron-independent cellular uptake
To Be Published
|
|
2F48
 
 | |
6LKG
 
 | | two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein, ... | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
