6WSL
 
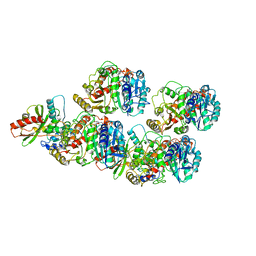 | | Cryo-EM structure of VASH1-SVBP bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Small vasohibin-binding protein, ... | | Authors: | Li, F, Li, Y, Yu, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of VASH1-SVBP bound to microtubules.
Elife, 9, 2020
|
|
6OCG
 
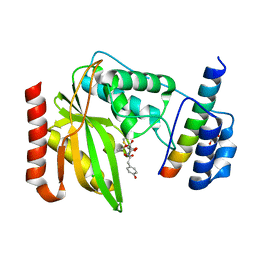 | | Crystal structure of VASH1-SVBP complex bound with EpoY | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[(3R)-4-ethoxy-3-hydroxy-4-oxobutanoyl]-L-tyrosine, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OCF
 
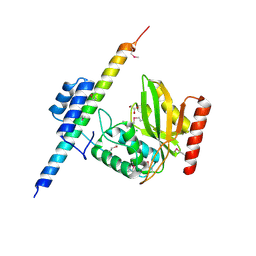 | | The crystal structure of VASH1-SVBP complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OCH
 
 | | Crystal structure of VASH1-SVBP complex bound with parthenolide | | Descriptor: | GLYCEROL, SULFATE ION, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2AJF
 
 | | Structure of SARS coronavirus spike receptor-binding domain complexed with its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme-Related Carboxypeptidase (Ace2), CHLORIDE ION, ... | | Authors: | Li, F, Li, W, Farzan, M, Harrison, S.C. | | Deposit date: | 2005-08-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of SARS coronavirus spike receptor-binding domain complexed with receptor.
Science, 309, 2005
|
|
8EON
 
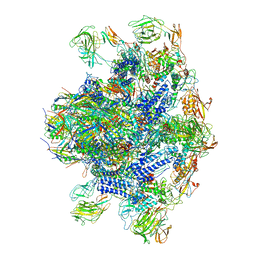 | | Pseudomonas phage E217 baseplate complex | | Descriptor: | Baseplate component gp33, Baseplate component gp34, Baseplate component gp36, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
8ENV
 
 | | In situ cryo-EM structure of Pseudomonas phage E217 tail baseplate in C6 map | | Descriptor: | Baseplate_J domain-containing protein gp44, Ripcord gp36, Sheath initiator gp34, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
8FRS
 
 | |
8FVG
 
 | |
8FVH
 
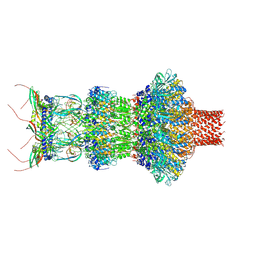 | | Pseudomonas phage E217 neck (portal, head-to-tail connector, collar and gateway proteins) | | Descriptor: | E217 collar protein gp28, E217 gateway protein gp29, E217 head-to-tail connector protein gp27, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
4XYW
 
 | | Glycosyltransferases WbnH | | Descriptor: | O-antigen biosynthesis glycosyltransferase WbnH, SULFATE ION | | Authors: | Li, F. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glycosyltransferases WbnH
To Be Published
|
|
8FUV
 
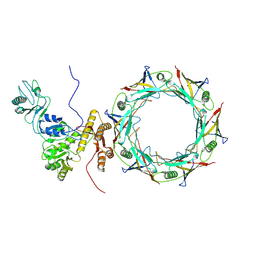 | |
4Y3I
 
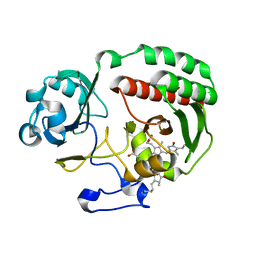 | | PAS-GAF fragment from Deinococcus radiodurans BphP assembled with BV - Y307S, low dose | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, F, Burgie, E.S, Yu, T, Heroux, A, Schatz, G.C, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-20 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
J.Am.Chem.Soc., 137, 2015
|
|
4Y5F
 
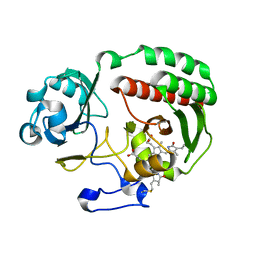 | | PAS-GAF fragment from Deinococcus radiodurans BphP assembled with BV - Y307S, high dose | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, F, Burgie, E.S, Yu, T, Heroux, A, Schatz, G.C, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
J.Am.Chem.Soc., 137, 2015
|
|
3UIU
 
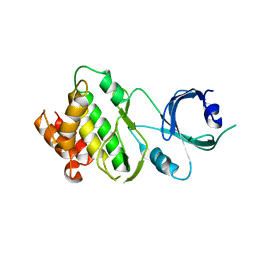 | | Crystal structure of Apo-PKR kinase domain | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
3D0H
 
 | |
3D0I
 
 | |
3D0G
 
 | |
1MIV
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme | | Descriptor: | MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
4ROU
 
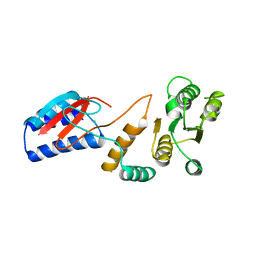 | |
4UC3
 
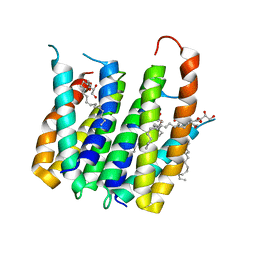 | | Translocator protein 18 kDa (TSPO) from Rhodobacter sphaeroides wild type | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
4UC1
 
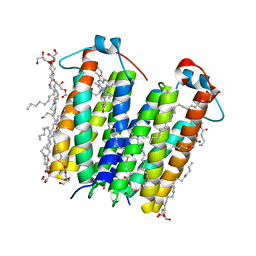 | | High resolution crystal structure of translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, METHOXY-ETHOXYL, ... | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
4UC2
 
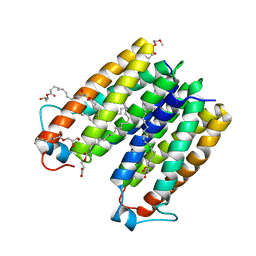 | | Crystal structure of translocator protein 18kDa (TSPO) from rhodobacter sphaeroides (A139T mutant) in P212121 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, TETRAETHYLENE GLYCOL, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
