1AVP
 
 | |
6RNT
 
 | | CRYSTAL STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH ADENOSINE 2'-MONOPHOSPHATE AT 1.8-ANGSTROMS RESOLUTION | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Ding, J, Koellner, G, Grunert, H.-P, Saenger, W. | | Deposit date: | 1991-08-20 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease T1 complexed with adenosine 2'-monophosphate at 1.8-A resolution.
J.Biol.Chem., 266, 1991
|
|
1HNI
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN A COMPLEX WITH THE NONNUCLEOSIDE INHIBITOR ALPHA-APA R 95845 AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Ding, J, Das, K, Arnold, E. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution.
Structure, 3, 1995
|
|
3A0E
 
 | | Crystal Structure of Polygonatum cyrtonema lectin (PCL) complexed with dimannoside | | Descriptor: | Mannose/sialic acid-binding lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2009-03-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a novel anti-HIV mannose-binding lectin from Polygonatum cyrtonema Hua with unique ligand-binding property and super-structure
J.Struct.Biol., 171, 2010
|
|
3A0C
 
 | |
2HMI
 
 | | HIV-1 REVERSE TRANSCRIPTASE/FRAGMENT OF FAB 28/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3'), FAB FRAGMENT OF MONOCLONAL ANTIBODY 28, ... | | Authors: | Ding, J, Arnold, E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution.
J.Mol.Biol., 284, 1998
|
|
2WEB
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL(2S)-[1-(((N-FORMYL)-L-VALYL)AMINO-2-(2-NAPHTHYL)ETHYL)HYDROXYPHOSPHINYLOXY]-3-PHENYLPROPANOATE, SODIUM SALT | | Descriptor: | METHYL (2S)-[1-((N-FORMYL)-L-VALYL)AMINO-2-(2-NAPHTHYL)ETHYL)HYDROXYPHOSPHINYLOXY]-3-PHENYL PROPANOATE, PENICILLOPEPSIN, SULFATE ION, ... | | Authors: | Ding, J, Fraser, M.E, James, M.N.G. | | Deposit date: | 1998-02-03 | | Release date: | 1998-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Macrocyclic Inhibitors of Penicillopepsin. II. X-Ray Crystallographic Analyses of Penicillopepsin Complexed with a P3-P1 Macrocyclic Peptidyl Inhibitor and with its Two Acyclic Analogues
J.Am.Chem.Soc., 120, 1998
|
|
2WEC
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL(2S)-[1-(((N-(1-NAPHTHALENEACETYL))-L-VALYL)AMINOMETHYL)HYDROXY PHOSPHINYLOXY]-3-PHENYLPROPANOATE, SODIUM SALT | | Descriptor: | METHYL (2S)-[1-((N-(NAPHTHALENEACETYL))-L-VALYL)AMINOMETHYL)HYDROXYPHOSPHINYLOXY]-3-PHENYL PROPANOATE, PENICILLOPEPSIN, SULFATE ION, ... | | Authors: | Ding, J, Fraser, M.E, James, M.N.G. | | Deposit date: | 1998-02-03 | | Release date: | 1998-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Macrocyclic Inhibitors of Penicillopepsin. II. X-Ray Crystallographic Analyses of Penicillopepsin Complexed with a P3-P1 Macrocyclic Peptidyl Inhibitor and with its Two Acyclic Analogues
J.Am.Chem.Soc., 120, 1998
|
|
2WEA
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL[CYCLO-7[(2R)-((N-VALYL) AMINO)-2-(HYDROXYL-(1S)-1-METHYOXYCARBONYL-2-PHENYLETHOXY) PHOSPHINYLOXY-ETHYL]-1-NAPHTHALENEACETAMIDE], SODIUM SALT | | Descriptor: | METHYL[CYCLO-7[(2R)-((N-VALYL)AMINO)-2-(HYDROXYL-(1S)-1-METHYLOXYCARBONYL-2-PHENYLETHOXY)PHOSPHINYLOXY-ETHYL]-1-NAPHTHALENEACETAMIDE], PENICILLOPEPSIN, SULFATE ION, ... | | Authors: | Ding, J, Fraser, M.E, James, M.N.G. | | Deposit date: | 1998-02-03 | | Release date: | 1998-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Macrocyclic Inhibitors of Penicillopepsin. II. X-Ray Crystallographic Analyses of Penicillopepsin Complexed with a P3-P1 Macrocyclic Peptidyl Inhibitor and with its Two Acyclic Analogues
J.Am.Chem.Soc., 120, 1998
|
|
2WED
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE MACROCYCLIC INHIBITOR:METHYL[CYCLO-7[(2R)-((N-VALYL)AMINO)-2-(HYDROXYL-(1S)-1-METHYOXYCARBONYL-2-PHENYLETHOXY)PHOSPHINYLOXY-ETHYL]-1-NAPHTHALENEACETAMIDE], SODIUM SALT | | Descriptor: | METHYL[CYCLO-7[(2R)-((N-VALYL)AMINO)-2-(HYDROXYL-(1S)-1-METHYLOXYCARBONYL-2-PHENYLETHOXY)PHOSPHINYLOXY-ETHYL]-1-NAPHTHALENEACETAMIDE], PENICILLOPEPSIN, SULFATE ION, ... | | Authors: | Ding, J, Fraser, M.E, James, M.N.G. | | Deposit date: | 1998-02-03 | | Release date: | 1998-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Macrocyclic Inhibitors of Penicillopepsin. II. X-Ray Crystallographic Analyses of Penicillopepsin Complexed with a P3-P1 Macrocyclic Peptidyl Inhibitor and with its Two Acyclic Analogues
J.Am.Chem.Soc., 120, 1998
|
|
5B5R
 
 | | Crystal structure of GSDMA3 | | Descriptor: | Gasdermin-A3 | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-15 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Pore-forming activity and structural autoinhibition of the gasdermin family.
Nature, 535, 2016
|
|
8RNT
 
 | | STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH ZINC(II) AT 1.8 ANGSTROMS RESOLUTION: A ZN2+.6H2O.CARBOXYLATE CLATHRATE | | Descriptor: | RIBONUCLEASE T1, ZINC ION | | Authors: | Ding, J, Choe, H.-W, Granzin, J, Saenger, W. | | Deposit date: | 1991-09-23 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ribonuclease T1 complexed with zinc(II) at 1.8 A resolution: a Zn2+.6H2O.carboxylate clathrate.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
3A0D
 
 | |
1K5M
 
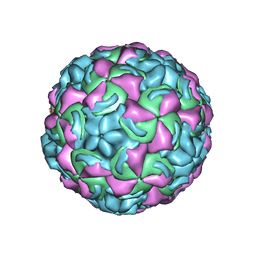 | | Crystal Structure of a Human Rhinovirus Type 14:Human Immunodeficiency Virus Type 1 V3 Loop Chimeric Virus MN-III-2 | | Descriptor: | CHIMERA OF HRV14 COAT PROTEIN VP2 (P1B) AND the V3 loop of HIV-1 gp120, COAT PROTEIN VP1 (P1D), COAT PROTEIN VP3 (P1C), ... | | Authors: | Ding, J, Smith, A.D, Geisler, S.C, Ma, X, Arnold, G.F, Arnold, E. | | Deposit date: | 2001-10-11 | | Release date: | 2002-07-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Human Rhinovirus
that Displays Part of the HIV-1 V3 Loop and
Induces Neutralizing Antibodies against
HIV-1
Structure, 10, 2002
|
|
3VPI
 
 | |
3VPJ
 
 | |
3WLD
 
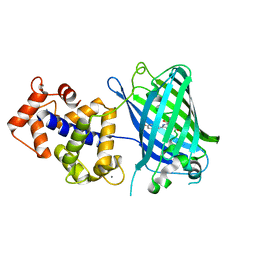 | | Crystal structure of monomeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
3WLC
 
 | | Crystal structure of dimeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
3AJM
 
 | | Crystal structure of programmed cell death 10 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Programmed cell death protein 10 | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human programmed cell death 10 complexed with inositol-(1,3,4,5)-tetrakisphosphate: a novel adaptor protein involved in human cerebral cavernous malformation.
Biochem.Biophys.Res.Commun., 399, 2010
|
|
2UP1
 
 | | STRUCTURE OF UP1-TELOMERIC DNA COMPLEX | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1) | | Authors: | Ding, J, Hayashi, M.K, Krainer, A.R, Xu, R.-M. | | Deposit date: | 1998-07-10 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA.
Genes Dev., 13, 1999
|
|
6ACI
 
 | | Crystal structure of EPEC effector NleB in complex with FADD death domain | | Descriptor: | FAS-associated death domain protein, MANGANESE (II) ION, T3SS secreted effector NleB homolog, ... | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-26 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
6AC0
 
 | | Crystal structure of TRADD death domain GlcNAcylated by EPEC effector NleB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
6AC5
 
 | | Crystal structure of RIPK1 death domain GlcNAcylated by EPEC effector NleB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-25 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
8SA4
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
