2VIJ
 
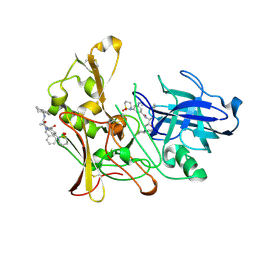 | | Human BACE-1 in complex with 3-(1,1-dioxidotetrahydro-2H-1,2-thiazin- 2-yl)-5-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(1,2,3,4- tetrahydro-1-naphthalenylamino)propyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(1S)-1,2,3,4-tetrahydronaphthalen-1-ylamino]propyl}-3-(1,1-dioxido-1,2-thiazinan-2-yl)-5-(ethylamino)benzamide | | Authors: | Beswick, P, Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Gleave, R, Hawkins, J, Hussain, I, Johnson, C.N, Macpherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Skidmore, J, Soleil, V, Smith, K.J, Stanway, S, Stemp, G, Stuart, A, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-04 | | Release date: | 2008-01-29 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 3: Identification of Hydroxy Ethylamines (Heas) with Nanomolar Potency in Cells.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VJ7
 
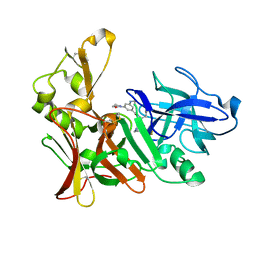 | | Human BACE-1 in complex with 3-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)-5-(2-oxo-1-pyrrolidinyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VJ9
 
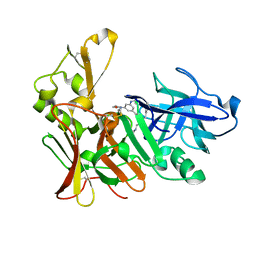 | | Human BACE-1 in complex with N-((1S,2R)-3-(cyclohexylamino)-2-hydroxy- 1-(phenylmethyl)propyl)-3-(ethylamino)-5-(2-oxo-1-pyrrolidinyl) benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-(cyclohexylamino)-2-hydroxypropyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-07 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VIZ
 
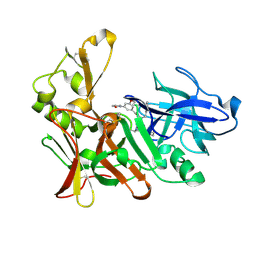 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-(2-oxo- 1-pyrrolidinyl)-5-(propyloxy)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-{[(1S)-2-(cyclohexylamino)-1-methyl-2-oxoethyl]amino}-2-hydroxypropyl]-3-(2-oxo-2,3-dihydro-1H-pyrrol-1-yl)-5-propoxybenzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 1: Identification of Novel Hydroxy Ethylamines (Heas).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VJ6
 
 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-(ethylamino)-5-(2-oxo-1-pyrrolidinyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-{[(1S)-2-(cyclohexylamino)-1-methyl-2-oxoethyl]amino}-2-hydroxypropyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VIY
 
 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-(pentylsulfonyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-{[(1S)-2-(cyclohexylamino)-1-methyl-2-oxoethyl]amino}-2-hydroxypropyl]-3-(pentylsulfonyl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Bace-1 Inhibitors Part 1: Identification of Novel Hydroxy Ethylamines (Heas).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1XSF
 
 | | Solution structure of a resuscitation promoting factor domain from Mycobacterium tuberculosis | | Descriptor: | Probable resuscitation-promoting factor rpfB | | Authors: | Cohen-Gonsaud, M, Barthe, P, Henderson, B, Ward, J, Roumestand, C, Keep, N.H. | | Deposit date: | 2004-10-19 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes
Nat.Struct.Mol.Biol., 12, 2005
|
|
2VIE
 
 | | Human BACE-1 in complex with N-((1S,2R)-1-benzyl-2-hydroxy-3-((1,1,5- trimethylhexyl)amino)propyl)-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl) benzamide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(1,1,5-trimethylhexyl)amino]propyl}-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3ZRR
 
 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CALCIUM ION, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ZRP
 
 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ZRQ
 
 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8P0D
 
 | | Human 14-3-3 sigma in complex with human MDM2 peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roversi, P, Ward, J, Doveston, R, Kwon, H, Romartinez Alonso, B. | | Deposit date: | 2023-05-10 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Characterizing the protein-protein interaction between MDM2 and 14-3-3 sigma ; proof of concept for small molecule stabilization.
J.Biol.Chem., 300, 2024
|
|
4QFR
 
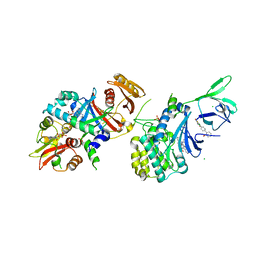 | | Structure of AMPK in complex with Cl-A769662 activator and STAUROSPORINE inhibitor | | Descriptor: | 2-chloro-4-hydroxy-3-(2'-hydroxybiphenyl-4-yl)-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
4QFS
 
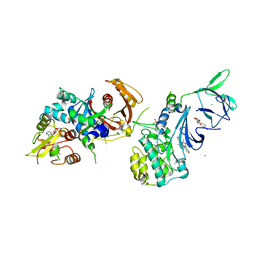 | | Structure of AMPK in complex with Br2-A769662core activator and STAUROSPORINE inhibitor | | Descriptor: | 2-bromo-3-(4-bromophenyl)-4-hydroxy-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
4CMF
 
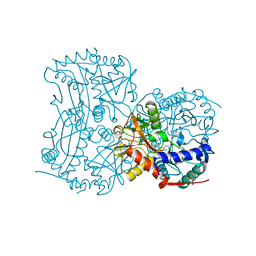 | | The (R)-selective transaminase from Nectria haematococca with inhibitor bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, AMINOTRANSFERASE, ... | | Authors: | Sayer, C, Isupov, M, Martinez-Torres, R.J, Richter, N, Hailes, H.C, Ward, J, Littlechild, J. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine:Pyruvate Transaminase from Nectria Haematococca.
FEBS J., 281, 2014
|
|
4CMD
 
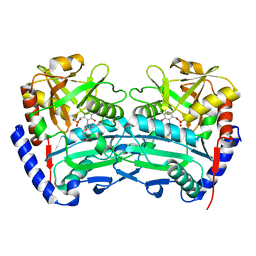 | | The (R)-selective transaminase from Nectria haematococca | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sayer, C, Isupov, M, Martinez-Torres, R.J, Richter, N, Hailes, H.C, Ward, J, Littlechild, J. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine:Pyruvate Transaminase from Nectria Haematococca.
FEBS J., 281, 2014
|
|
5KQ5
 
 | | AMPK bound to allosteric activator | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2016-07-05 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Discovery and Preclinical Characterization of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carboxylic Acid (PF-06409577), a Direct Activator of Adenosine Monophosphate-activated Protein Kinase (AMPK), for the Potential Treatment of Diabetic Nephropathy.
J.Med.Chem., 59, 2016
|
|
8EGU
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2~{S})-3-methyl-2-[pentanoyl-[[4-[2-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]amino]butanoic acid, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGF
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | (5P)-5-(4'-methyl[1,1'-biphenyl]-2-yl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, POTASSIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGD
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | 5-(4-methoxyphenyl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGQ
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2S)-2-ethyl-4-{[(2'M)-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
6HT1
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with SGC-iMLLT (compound 92) | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6E4T
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-{6-chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carbonyl}-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
6E4U
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-{6-chloro-5-[6-(dimethylamino)-2-methoxypyridin-3-yl]-1H-indole-3-carbonyl}-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
