8E8W
 
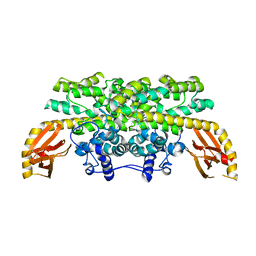 | |
6VZY
 
 | |
6XCV
 
 | |
6DQX
 
 | | Actinobacillus ureae class Id ribonucleotide reductase alpha subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | McBride, M.J, Palowitch, G.M, Boal, A.K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Class Id Ribonucleotide Reductase Catalytic Subunits Reveal a Minimal Architecture for Deoxynucleotide Biosynthesis.
Biochemistry, 58, 2019
|
|
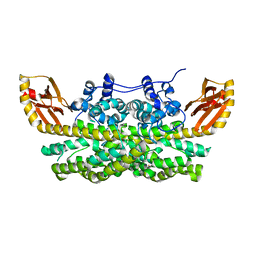
7TWA
 
 | | Crystal structure of apo BesC from Streptomyces cattleya | | Descriptor: | 1,3-BUTANEDIOL, 4-chloro-allylglycine synthase, ACETATE ION, ... | | Authors: | Neugebauer, M.E, McBride, M.J, Boal, A.K, Chang, M.C.Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate-Triggered mu-Peroxodiiron(III) Intermediate in the 4-Chloro-l-Lysine-Fragmenting Heme-Oxygenase-like Diiron Oxidase (HDO) BesC: Substrate Dissociation from, and C4 Targeting by, the Intermediate.
Biochemistry, 61, 2022
|
|
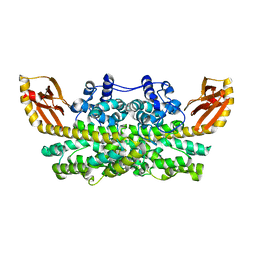
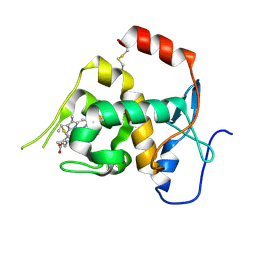
6ONQ
 
 | |
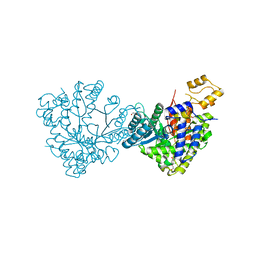
6DQW
 
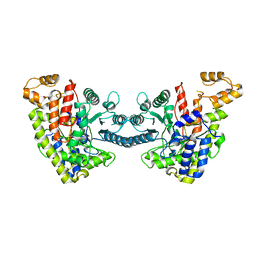 | |
5OLC
 
 | |
5OPQ
 
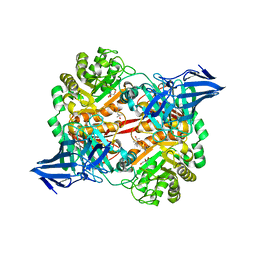 | | A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-D-galactosidase, ... | | Authors: | Ficko-Blean, E, Michel, G, Czjzek, M. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.
Nat Commun, 8, 2017
|
|
5IHS
 
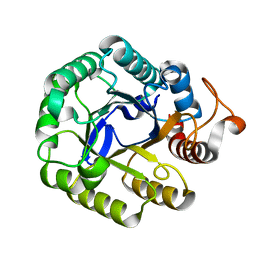 | | Structure of CHU_2103 from Cytophaga hutchinsonii | | Descriptor: | Endoglucanase, glycoside hydrolase family 5 protein | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Periplasmic Cytophaga hutchinsonii Endoglucanases Are Required for Use of Crystalline Cellulose as the Sole Source of Carbon and Energy.
Appl.Environ.Microbiol., 82, 2016
|
|
5J5U
 
 | | Fjoh_4561 chitin-binding protein | | Descriptor: | RagB/SusD domain protein | | Authors: | Koropatkin, N.M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin.
Biotechnol Biofuels, 9, 2016
|
|
5J90
 
 | |
6ONP
 
 | |
