1E5D
 
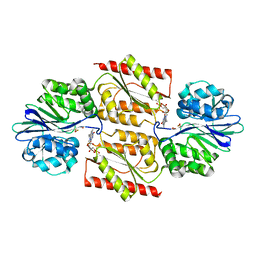 | | RUBREDOXIN OXYGEN:OXIDOREDUCTASE (ROO) FROM ANAEROBE DESULFOVIBRIO GIGAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, OXYGEN MOLECULE, ... | | Authors: | Frazao, C, Silva, G, Gomes, C.M, Matias, P, Coelho, R, Sieker, L, Macedo, S, Liu, M.Y, Oliveira, S, Teixeira, M, Xavier, A.V, Rodrigues-Pousada, C, Carrondo, M.A, Le Gall, J. | | Deposit date: | 2000-07-24 | | Release date: | 2000-11-17 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Nat.Struct.Biol., 7, 2000
|
|
3BH4
 
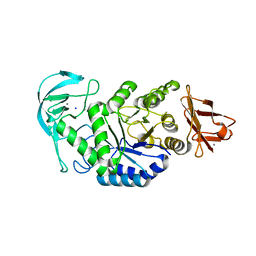 | | High resolution crystal structure of Bacillus amyloliquefaciens alpha-amylase | | Descriptor: | Alpha-amylase, CALCIUM ION, SODIUM ION | | Authors: | Alikhajeh, J, Khajeh, K, Ranjbar, B, Naderi-Manesh, H, Lin, Y.H, Liu, M.Y, Chen, C.J. | | Deposit date: | 2007-11-28 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Bacillus amyloliquefaciens alpha-amylase at high resolution: implications for thermal stability.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1KCB
 
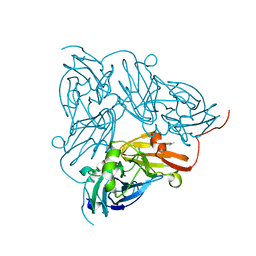 | | Crystal Structure of a NO-forming Nitrite Reductase Mutant: an Analog of a Transition State in Enzymatic Reaction | | Descriptor: | COPPER (II) ION, Nitrite Reductase | | Authors: | Liu, S.Q, Chang, T, Liu, M.Y, LeGall, J, Chang, W.C, Zhang, J.P, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-11-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a NO-forming nitrite reductase mutant: an analog of a transition state in enzymatic reaction
Biochem.Biophys.Res.Commun., 302, 2003
|
|
1DVB
 
 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, RUBRERYTHRIN, ZINC ION | | Authors: | Sieker, L.C, Holmes, M, Le Trong, I, Turley, S, Liu, M.Y, Legall, J, Stenkamp, R.E. | | Deposit date: | 2000-01-20 | | Release date: | 2000-11-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A crystal structure of the "as isolated" rubrerythrin from Desulfovibrio vulgaris: some surprising results.
J.Biol.Inorg.Chem., 5, 2000
|
|
1GNL
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans X-ray structure at 1.25A resolution using synchrotron radiation at a wavelength of 0.933A | | Descriptor: | ACETATE ION, HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1GN9
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans ATCC 27774 X-ray structure at 2.6A resolution using synchrotron radiation at a wavelength of 1.722A | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-04 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1GNT
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris. X-ray structure at 1.25A resolution using synchrotron radiation. | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 7, 2002
|
|
2AVF
 
 | | Crystal Structure of C-terminal Desundecapeptide Nitrite Reductase from Achromobacter cycloclastes | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Li, H.T, Chang, T, Chang, W.C, Chen, C.J, Liu, M.Y, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2005-08-30 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal desundecapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 338, 2005
|
|
3OR2
 
 | | Crystal structure of dissimilatory sulfite reductase II (DsrII) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissimilatory Sulfite Reductase, Sulfate Reduction
To be Published
|
|
3OR1
 
 | | Crystal structure of dissimilatory sulfite reductase I (DsrI) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure insights into the enzyme catalysis from comparison of three forms of dissimilatory sulfite reductase from Desulfovibrio gigas
To be Published
|
|
1JYB
 
 | | Crystal structure of Rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Chang, W.R, Li, M, Liu, M.Y. | | Deposit date: | 2001-09-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure studies on rubrerythrin: enzymatic activity in relation to the zinc movement.
J.Biol.Inorg.Chem., 8, 2003
|
|
1JR9
 
 | | Crystal Structure of manganese superoxide dismutases from Bacillus halodenitrificans | | Descriptor: | MANGANESE (II) ION, ZINC ION, manganese superoxide dismutase | | Authors: | Liao, J, Liu, M.Y, Chang, T, Li, M, LeGall, J, Gui, L.L, Zhang, J.P, Jiang, T, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-08-13 | | Release date: | 2002-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of manganese superoxide dismutase from Bacillus halodenitrificans, a component of the so-called "green protein".
J.Struct.Biol., 139, 2002
|
|
1NF6
 
 | | X-ray structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different catalytic states ("cycled" structure: reduced in solution and allowed to reoxidise before crystallisation) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (III) ION, GLYCEROL, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NF4
 
 | | X-Ray Structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different states (reduced structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (II) ION, SULFATE ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NFV
 
 | | X-ray structure of Desulfovibrio desulfuricans bacterioferritin: the diiron centre in different catalytic states (as-isolated structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, 3-HYDROXYPYRUVIC ACID, FE (III) ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
2GJ5
 
 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
1OA1
 
 | | REDUCED HYBRID CLUSTER PROTEIN (HCP) FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH STRUCTURE AT 1.55A RESOLUTION USING SYNCHROTRON RADIATION. | | Descriptor: | FE4-S3 CLUSTER, GLYCEROL, HYDROXYLAMINE REDUCTASE, ... | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Romao, C.V, Liu, M.Y, Frazao, C, Saraiva, L.M, Xavier, A.V, Legall, J, Van Dongen, W.M.A.M, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P.F. | | Deposit date: | 2002-12-23 | | Release date: | 2003-04-08 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Reduced Hybrid Cluster Proteins (Hcp) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at High Resolution Using Synchrotron Radiation
J.Biol.Inorg.Chem., 8, 2003
|
|
1OA0
 
 | | REDUCED HYBRID CLUSTER PROTEIN FROM DESULFOVIBRIO DESULFURICANS X-RAY STRUCTURE AT 1.25A RESOLUTION | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE4-S3 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Macedo, S, Aragao, D, Mitchell, E.P, Romao, C.V, Liu, M.Y, Frazao, C, Saraiva, L.M, Xavier, A.V, Legall, J, Van Dongen, W.M.A.M, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P.F. | | Deposit date: | 2002-12-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reduced hybrid cluster proteins (HCP) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at high resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 8, 2003
|
|
3UYY
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UZO
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UZB
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
1RZQ
 
 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH5.0 | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2003-12-26 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1RZP
 
 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH6.2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2003-12-26 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1B71
 
 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, PROTEIN (RUBRERYTHRIN), ZINC ION | | Authors: | Sieker, L.C, Holmes, M, Le Trong, I, Turley, S, Santarsiero, B.D, Liu, M.-Y, Legall, J, Stenkamp, R.E. | | Deposit date: | 1999-01-26 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative metal-binding sites in rubrerythrin.
Nat.Struct.Biol., 6, 1999
|
|
1Z28
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Phenol-sulfating phenol sulfotransferase 1 | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
