2Z9L
 
 | |
2ZU3
 
 | | Complex structure of CVB3 3C protease with TG-0204998 | | Descriptor: | 3C proteinase, N-[(benzyloxy)carbonyl]-3-[(2,2-dimethylpropanoyl)amino]-L-alanyl-N-[(1R)-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}pentyl]-L-leucinamide | | Authors: | Lee, C.C, Tsui, Y.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2Z9G
 
 | | Complex structure of SARS-CoV 3C-like protease with PMA | | Descriptor: | 3C-like proteinase, BENZENE, MERCURY (II) ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
2ZTZ
 
 | |
2Z94
 
 | | Complex structure of SARS-CoV 3C-like protease with TDT | | Descriptor: | 4-methylbenzene-1,2-dithiol, Replicase polyprotein 1ab, ZINC ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-17 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors
Febs Lett., 581, 2007
|
|
2Z9K
 
 | | Complex structure of SARS-CoV 3C-like protease with JMF1600 | | Descriptor: | (dimethylamino)(hydroxy)zinc', 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
2ZU2
 
 | | complex structure of CoV 229E 3CL protease with EPDTC | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZTY
 
 | |
2ZTX
 
 | | Complex structure of CVB3 3C protease with EPDTC | | Descriptor: | 3C proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2Z9J
 
 | | Complex structure of SARS-CoV 3C-like protease with EPDTC | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
2ZU1
 
 | |
3LCT
 
 | |
3LCS
 
 | |
1J7J
 
 | |
1KEX
 
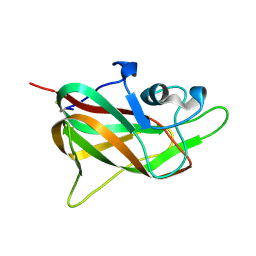 | | Crystal Structure of the b1 Domain of Human Neuropilin-1 | | Descriptor: | Neuropilin-1 | | Authors: | Lee, C.C, Kreusch, A, McMullan, D, Ng, K, Spraggon, G. | | Deposit date: | 2001-11-18 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Neuropilin-1 b1 Domain
Structure, 11, 2003
|
|
3WUR
 
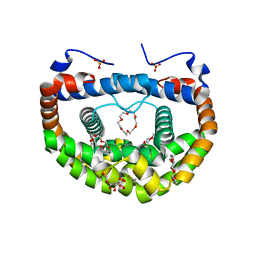 | | Structure of DMP19 Complex with 18-crown-6 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, L(+)-TARTARIC ACID, ... | | Authors: | Lee, C.C, Wang, H.C, Wang, A.H.J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WH0
 
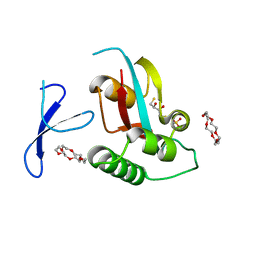 | | Structure of Pin1 Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Lee, C.C, Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WHM
 
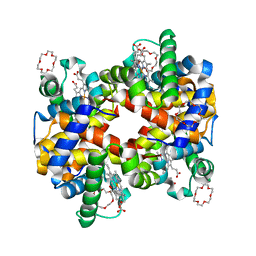 | | Structure of Hemoglobin Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Lee, C.C, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1NH1
 
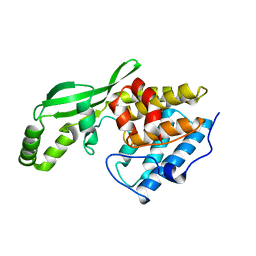 | | Crystal Structure of the Type III Effector AvrB from Pseudomonas syringae. | | Descriptor: | Avirulence B protein | | Authors: | Lee, C.C, Wood, M.D, Ng, K, Luginbuhl, P, Spraggon, G, Katagiri, F. | | Deposit date: | 2002-12-18 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Type III Effector AvrB from Pseudomonas syringae.
Structure, 12, 2004
|
|
1ZLT
 
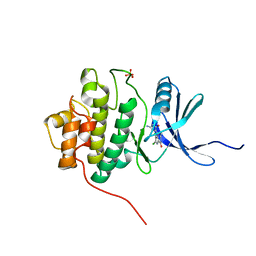 | | Crystal Structure of Chk1 Complexed with a Hymenaldisine Analog | | Descriptor: | (4Z)-4-(2-AMINO-5-OXO-3,5-DIHYDRO-4H-IMIDAZOL-4-YLIDENE)-2,3-DICHLORO-4,5,6,7-TETRAHYDROPYRROLO[2,3-C]AZEPIN-8(1H)-ONE, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Lee, C.C, Ng, K, Wan, Y, Gray, N, Spraggon, G. | | Deposit date: | 2005-05-09 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Chk1 Complexed with a Hymenaldisine Analog
To be Published
|
|
3Q6N
 
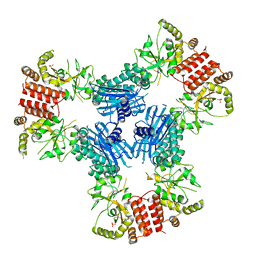 | | Crystal Structure of Human MC-HSP90 in P21 space group | | Descriptor: | Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Lee, C.C, Lin, T.W, Ko, T.P, Wang, A.H.-J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The hexameric structures of human heat shock protein 90
Plos One, 6, 2011
|
|
3Q6M
 
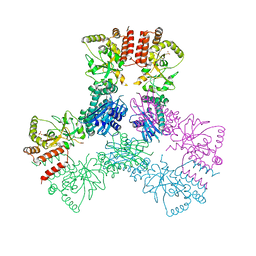 | | Crystal Structure of Human MC-HSP90 in C2221 Space Group | | Descriptor: | Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Lee, C.C, Lin, T.W, Ko, T.P, Wang, A.H.-J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The hexameric structures of human heat shock protein 90
Plos One, 6, 2011
|
|
4XZB
 
 | |
3W9D
 
 | |
3W9E
 
 | | Structure of Human Monoclonal Antibody E317 Fab Complex with HSV-2 gD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab heavy chain, Antibody Fab light chain, ... | | Authors: | Lee, C.C, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the antibody neutralization of herpes simplex virus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
