3BB1
 
 | | Crystal structure of Toc34 from Pisum sativum in complex with Mg2+ and GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Koenig, P, Sinning, I, Schleiff, E, Tews, I. | | Deposit date: | 2007-11-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures.
Structure, 16, 2008
|
|
3MC9
 
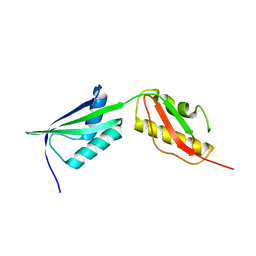 | |
3MC8
 
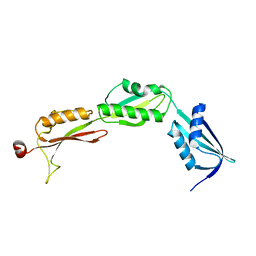 | |
3BB3
 
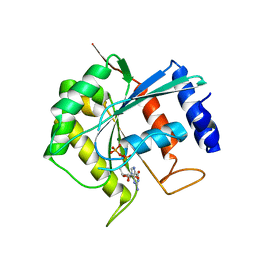 | | Crystal structure of Toc33 from Arabidopsis thaliana in complex with GDP and Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | Authors: | Koenig, P, Sinning, I, Schleiff, E, Tews, I. | | Deposit date: | 2007-11-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures.
Structure, 16, 2008
|
|
3BB4
 
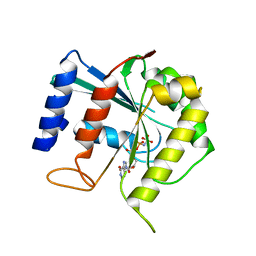 | | Crystal structure of Toc33 from Arabidopsis thaliana in complex with Mg2+ and GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, T7I23.11 protein | | Authors: | Koenig, P, Sinning, I, Schleiff, E, Tews, I. | | Deposit date: | 2007-11-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures.
Structure, 16, 2008
|
|
3DEF
 
 | | Crystal structure of Toc33 from Arabidopsis thaliana, dimerization deficient mutant R130A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | Authors: | Koenig, P, Schleiff, E, Sinning, I, Tews, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | On the significance of Toc-GTPase homodimers
J.Biol.Chem., 283, 2008
|
|
4EV1
 
 | | Anabaena Tic22 (protein transport) | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Anabena Tic22, ... | | Authors: | Koenig, P, Schleiff, E, Sinning, I, Tews, I. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional conservation of Tic22 in cyanobacterial outer membrane protein assembly and chloroplast translocation.
TO BE PUBLISHED
|
|
2DGC
 
 | |
1DGC
 
 | |
4ZFG
 
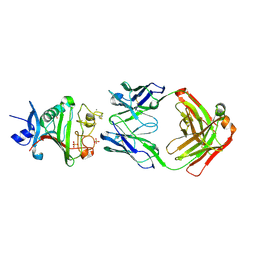 | | Dual-specificity Fab 5A12 in complex with Angiopoietin 2 | | Descriptor: | Angiopoietin-2, CALCIUM ION, Fragment antigen binding 5A12 heavy chain, ... | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Deep Sequencing-guided Design of a High Affinity Dual Specificity Antibody to Target Two Angiogenic Factors in Neovascular Age-related Macular Degeneration.
J.Biol.Chem., 290, 2015
|
|
4ZFF
 
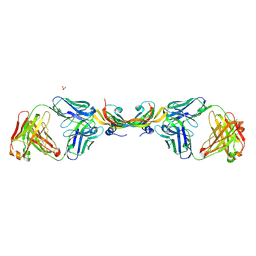 | | Dual-acting Fab 5A12 in complex with VEGF | | Descriptor: | Fragment antigen binding (Fab) 5A12 Heavy chain, Fragment antigen binding (Fab) 5A12 Light chain, SULFATE ION, ... | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Deep Sequencing-guided Design of a High Affinity Dual Specificity Antibody to Target Two Angiogenic Factors in Neovascular Age-related Macular Degeneration.
J.Biol.Chem., 290, 2015
|
|
1IGN
 
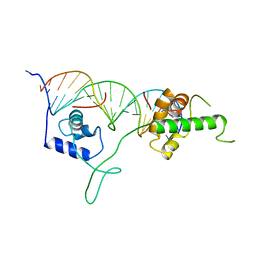 | | DNA-BINDING DOMAIN OF RAP1 IN COMPLEX WITH TELOMERIC DNA SITE | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*AP*CP*AP*CP*CP*CP*AP*CP*AP*CP*AP*CP*C P*AP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*GP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G P*CP*G)-3'), PROTEIN (RAP1) | | Authors: | Koenig, P, Giraldo, R, Chapman, L, Rhodes, D. | | Deposit date: | 1996-02-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA.
Cell(Cambridge,Mass.), 85, 1996
|
|
5GCH
 
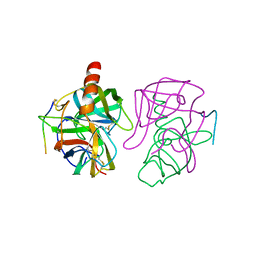 | |
6VGO
 
 | |
6VGR
 
 | |
8C51
 
 | | Trypanosoma brucei IMP dehydrogenase (cyto) crystallized in High Five cells revealing native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C5K
 
 | | HEX-1 (in cellulo, in situ) crystallized and diffracted in High Five cells. Growth and SX data collection at 296 K on CrystalDirect plates | | Descriptor: | Woronin body major protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD5
 
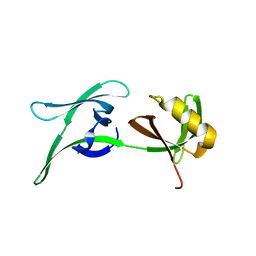 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD4
 
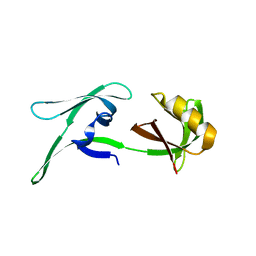 | | structure of HEX-1 from N. crassa crystallized in cellulo (cytosol), diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD6
 
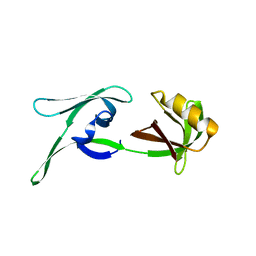 | | structure of HEX-1 (cyto V2) from N. crassa grown in living insect cells, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CGX
 
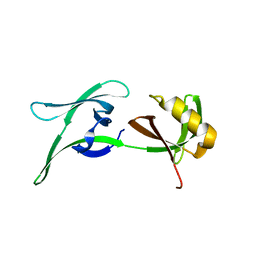 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using XDS | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
4GCH
 
 | |
6GCH
 
 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | Descriptor: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | Authors: | Brady, K, Wei, A, Ringe, D, Abeles, R.H. | | Deposit date: | 1990-04-06 | | Release date: | 1990-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
7GCH
 
 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | Descriptor: | 1,1,1-TRIFLUORO-3-((N-ACETYL)-L-LEUCYLAMIDO)-4-PHENYL-BUTAN-2-ONE(N-ACETYL-L-LEUCYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | Authors: | Brady, K, Ringe, D, Abeles, R.H. | | Deposit date: | 1990-04-06 | | Release date: | 1990-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
3GCH
 
 | |
