6IME
 
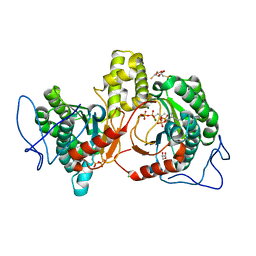 | | Rv2361c complex with substrate analogues | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, CARBONATE ION, ... | | Authors: | Ko, T.-P, Guo, R.-T, Chen, C.-C, Liu, W. | | Deposit date: | 2018-10-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate-analogue complex structure of Mycobacterium tuberculosis decaprenyl diphosphate synthase.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7EBD
 
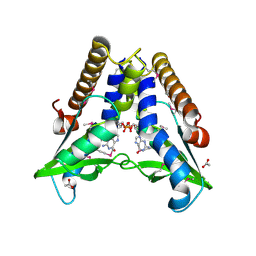 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, TIR-like domain-containing protein | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
7EBL
 
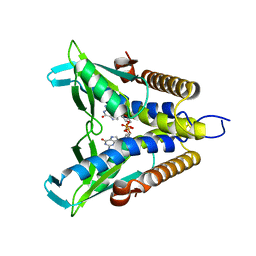 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), STING | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
1UAQ
 
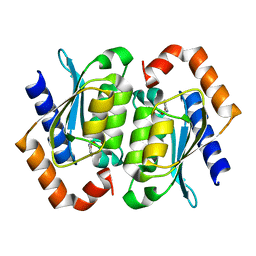 | | The crystal structure of yeast cytosine deaminase | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, ZINC ION, cytosine deaminase | | Authors: | Ko, T.-P, Lin, J.-J, Hu, C.-Y, Hsu, Y.-H, Wang, A.H.-J, Liaw, S.-H. | | Deposit date: | 2003-03-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution
J.Biol.Chem., 278, 2003
|
|
2CAU
 
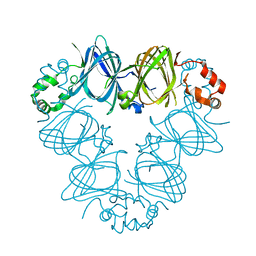 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2CAV
 
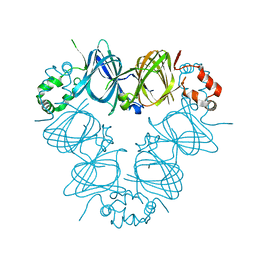 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DGR
 
 | |
1DGW
 
 | |
5GWV
 
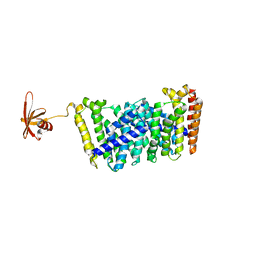 | | Structure of MoeN5-Sso7d fusion protein in complex with a substrate analogue | | Descriptor: | (2R)-3-dimethoxyphosphoryloxy-2-[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoic acid, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5GWW
 
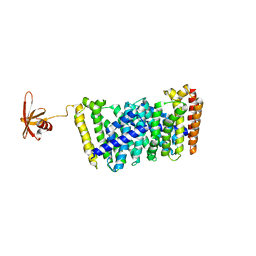 | | Structure of MoeN5-Sso7d fusion protein in complex with a permethylated substrate analogue | | Descriptor: | MoeN5,DNA-binding protein 7d, methyl (2R)-3-dimethoxyphosphoryloxy-2-[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoate | | Authors: | Ko, T.-P, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5B01
 
 | |
3LBM
 
 | | D-sialic acid aldolase | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Ko, T.-P, Chou, C.-Y, Wang, A.H.-J. | | Deposit date: | 2010-01-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
4EM2
 
 | |
1M7J
 
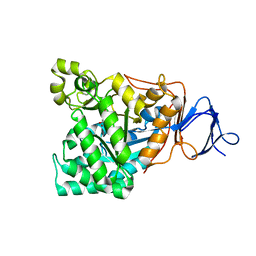 | | Crystal structure of D-aminoacylase defines a novel subset of amidohydrolases | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Liaw, S.-H, Chen, S.-J, Ko, T.-P, Hsu, C.-S, Wang, A.H.-J, Tsai, Y.-C. | | Deposit date: | 2002-07-22 | | Release date: | 2003-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of D-Aminoacylase from Alcaligenes faecalis DA1. A NOVEL SUBSET OF AMIDOHYDROLASES AND INSIGHTS INTO THE ENZYME MECHANISM.
J.Biol.Chem., 278, 2003
|
|
3FWN
 
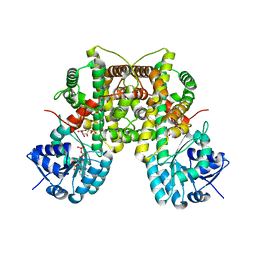 | | Dimeric 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
1XYI
 
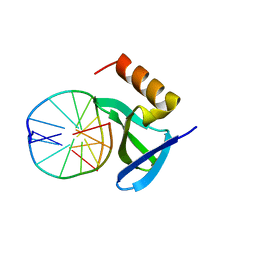 | | Hyperthermophile chromosomal protein Sac7d double mutant Val26Ala/Met29Ala in complex with DNA GCGATCGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-10 | | Release date: | 2005-02-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
Nucleic Acids Res., 33, 2005
|
|
2DH4
 
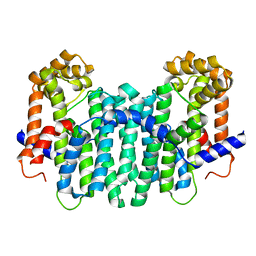 | | Geranylgeranyl pyrophosphate synthase | | Descriptor: | MAGNESIUM ION, YPL069C | | Authors: | Chang, T.-H, Guo, R.-T, Ko, T.-P, Wang, A.H, Liang, P.-H. | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of Type-III Geranylgeranyl Pyrophosphate Synthase from Saccharomyces cerevisiae and the Mechanism of Product Chain Length Determination.
J.Biol.Chem., 281, 2006
|
|
7XGS
 
 | | Short vegetative phase protein | | Descriptor: | 1,2-ETHANEDIOL, MADS-box protein SVP | | Authors: | Liao, Y.-T, Wang, H.-C, Ko, T.-P. | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The PHYL1 effector of peanut witches broom (PnWB) phytoplasma alters the miR156/157-mediated squamosa promoter binding protein like regulation and gibberellic acid generation to promote anthocyanin biosynthesis
To Be Published
|
|
1UEH
 
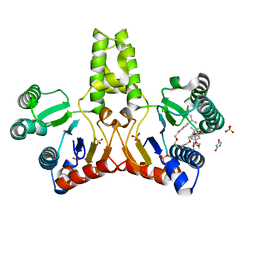 | | E. coli undecaprenyl pyrophosphate synthase in complex with Triton X-100, magnesium and sulfate | | Descriptor: | MAGNESIUM ION, OXTOXYNOL-10, SULFATE ION, ... | | Authors: | Chang, S.-Y, Ko, T.-P, Liang, P.-H, Wang, A.H.-J. | | Deposit date: | 2003-05-15 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton
J.Biol.Chem., 278, 2003
|
|
5FNN
 
 | | Iron and Selenomethionine containing Iron sulfur cluster repair protein YtfE | | Descriptor: | CHLORIDE ION, FE (II) ION, FE (III) ION, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-27 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
5FNP
 
 | | High resolution Zn containing Iron sulfur cluster repair protein YtfE | | Descriptor: | CHLORIDE ION, IRON-SULFUR CLUSTER REPAIR PROTEIN YTFE, OXYGEN ATOM, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
5FNY
 
 | | Low solvent content crystal form of Zn containing Iron sulfur cluster repair protein YtfE | | Descriptor: | FE (III) ION, IRON-SULFUR CLUSTER REPAIR PROTEIN YTFE, NONAETHYLENE GLYCOL, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
3W24
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 | | Descriptor: | Glycoside hydrolase family 10 | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W4U
 
 | | Human zeta-2 beta-2-s hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit beta, Hemoglobin subunit zeta, ... | | Authors: | Safo, M.K, Ko, T.-P, Russell, J.E. | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of fully liganded Hb zeta 2 beta 2(s) trapped in a tense conformation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WGH
 
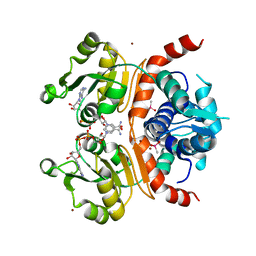 | | Crystal structure of RSP in complex with beta-NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CACODYLATE ION, Redox-sensing transcriptional repressor rex, ... | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
