2KZB
 
 | | Solution structure of alpha-mannosidase binding domain of Atg19 | | Descriptor: | Autophagy-related protein 19 | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
2KZK
 
 | | Solution structure of alpha-mannosidase binding domain of Atg34 | | Descriptor: | Uncharacterized protein YOL083W | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
4YTV
 
 | | Crystal structure of Mdm35 | | Descriptor: | COBALT (II) ION, GLYCEROL, Mitochondrial distribution and morphology protein 35 | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YTW
 
 | | Crystal structure of Ups1-Mdm35 complex | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YTX
 
 | | Crystal structure of Ups1-Mdm35 complex with PA | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
7BYU
 
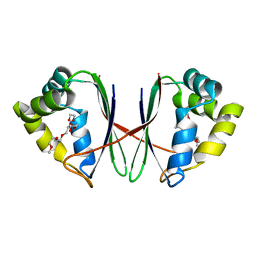 | | Crystal structure of Acidovorax avenae L-fucose mutarotase (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, L-fucose mutarotase | | Authors: | Watanabe, Y, Fukui, Y, Watanabe, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Functional and structural characterization of a novel L-fucose mutarotase involved in non-phosphorylative pathway of L-fucose metabolism.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
7BYW
 
 | |
6J7C
 
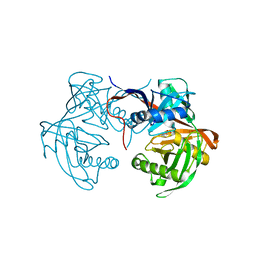 | | Crystal structure of proline racemase-like protein from Thermococcus litoralis in complex with proline | | Descriptor: | PROLINE, Proline racemase | | Authors: | Watanabe, Y, Watanabe, S, Itoh, Y, Watanabe, Y. | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of substrate-bound bifunctional proline racemase/hydroxyproline epimerase from a hyperthermophilic archaeon.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
3VU4
 
 | |
5AUJ
 
 | |
7YPD
 
 | |
6JNJ
 
 | |
6JNK
 
 | |
6L06
 
 | |
6L07
 
 | |
6K9Y
 
 | | Crystal structure of human VAT-1 | | Descriptor: | NITRATE ION, Synaptic vesicle membrane protein VAT-1 homolog | | Authors: | Watanabe, Y, Endo, T. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for interorganelle phospholipid transport mediated by VAT-1.
J.Biol.Chem., 295, 2020
|
|
5JGE
 
 | |
5AZH
 
 | | Crystal structure of LGG-2 fused with an EEEWEEL peptide | | Descriptor: | EEEWEEL peptide,Protein lgg-2, MAGNESIUM ION | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
5AZG
 
 | | Crystal structure of LGG-1 complexed with a UNC-51 peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, Serine/threonine-protein kinase unc-51 | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
5AON
 
 | | Crystal structure of the conserved N-terminal domain of Pex14 from Trypanosoma brucei | | Descriptor: | PEROXIN 14, SULFATE ION | | Authors: | Obita, T, Sugawara, Y, Mizuguchi, M, Watanabe, Y, Kawaguchi, K, Imanaka, T. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Characterization of the Interaction between Trypanosoma Brucei Pex5P and its Receptor Pex14P.
FEBS Lett., 590, 2016
|
|
5AZF
 
 | | Crystal structure of LGG-1 complexed with a WEEL peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, SULFATE ION, ... | | Authors: | Watanabe, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
7C0D
 
 | |
7C0E
 
 | |
7C0C
 
 | |
2E2Y
 
 | | Crystal Structure of F43W/H64D/V68I Myoglobin | | Descriptor: | GLYCEROL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ohki, T, Ueno, T, Hikage, T, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2006-11-18 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reactivities of oxo and peroxo intermediates studied by hemoprotein mutants
Acc.Chem.Res., 40, 2007
|
|
