4N40
 
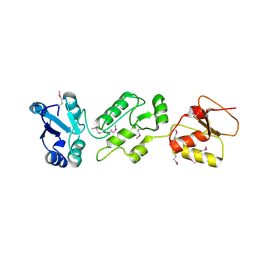 | | Crystal structure of human Epithelial cell-transforming sequence 2 protein | | Descriptor: | Protein ECT2 | | Authors: | Zou, Y, Shao, Z.H, Li, F.D, Gong, D, Wang, C, Gong, Q, Shi, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-08-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Crystal structure of triple-BRCT-domain of ECT2 and insights into the binding characteristics to CYK-4
Febs Lett., 588, 2014
|
|
2I8U
 
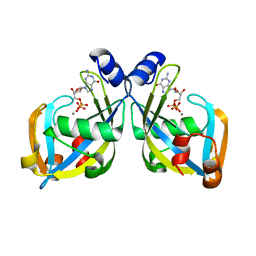 | | GDP-mannose mannosyl hydrolase-calcium-GDP product complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
2I8T
 
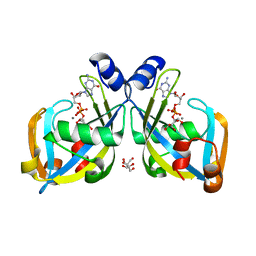 | | GDP-mannose mannosyl hydrolase-calcium-GDP-mannose complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GLYCEROL, ... | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
3DFI
 
 | |
3DFK
 
 | |
3DFF
 
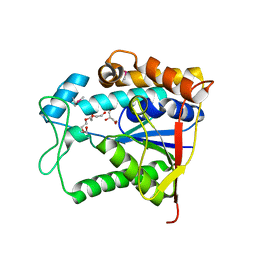 | | The crystal structure of teicoplanin pseudoaglycone deacetylase Orf2 | | Descriptor: | GLYCEROL, TETRAETHYLENE GLYCOL, Teicoplanin pseudoaglycone deacetylases Orf2, ... | | Authors: | Zou, Y, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-06-11 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of lipoglycopeptide antibiotic deacetylases: implications for the biosynthesis of a40926 and teicoplanin.
Chem.Biol., 15, 2008
|
|
3DFM
 
 | |
4E5P
 
 | |
4EBF
 
 | |
4E5K
 
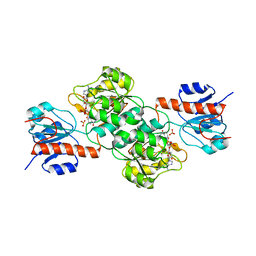 | |
4E5N
 
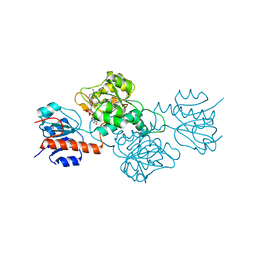 | | Thermostable phosphite dehydrogenase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Thermostable phosphite dehydrogenase | | Authors: | Zou, Y, Zhang, H, Nair, S.K. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of phosphite dehydrogenase provide insights into nicotinamide cofactor regeneration.
Biochemistry, 51, 2012
|
|
4E5M
 
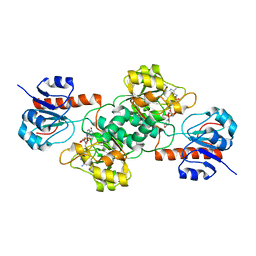 | |
4GBR
 
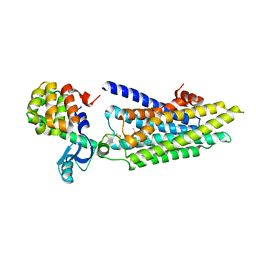 | |
3IX4
 
 | | LasR-TP1 complex | | Descriptor: | 2,4-dibromo-6-({[(2-nitrophenyl)carbonyl]amino}methyl)phenyl 2-chlorobenzoate, Transcriptional activator protein lasR | | Authors: | Zou, Y, Nair, S.K. | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
3IX8
 
 | | LasR-TP3 complex | | Descriptor: | 2,4-dibromo-6-({[(2-chlorophenyl)carbonyl]amino}methyl)phenyl 2-methylbenzoate, Transcriptional activator protein lasR | | Authors: | Zou, Y, Nair, S.K. | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
3IX3
 
 | | LasR-OC12 HSL complex | | Descriptor: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, Transcriptional activator protein lasR | | Authors: | Zou, Y, Nair, S.K. | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | LasR-OC12 HSL complex
TO BE PUBLISHED
|
|
3JPU
 
 | | LasR-TP4 complex | | Descriptor: | 4-bromo-2-({[(2-chlorophenyl)carbonyl]amino}methyl)-6-methylphenyl 2,4-dichlorobenzoate, Transcriptional activator protein lasR | | Authors: | Zou, Y, Nair, S.K. | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
2QKW
 
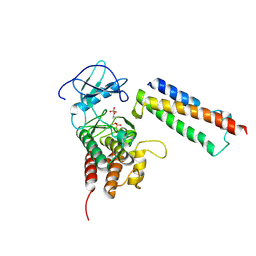 | | Structural basis for activation of plant immunity by bacterial effector protein AvrPto | | Descriptor: | Avirulence protein, Protein kinase | | Authors: | Xing, W.M, Zou, Y, Liu, Q, Hao, Q, Zhou, J.M, Chai, J.J. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis for activation of plant immunity by bacterial effector protein AvrPto
Nature, 449, 2007
|
|
3SN6
 
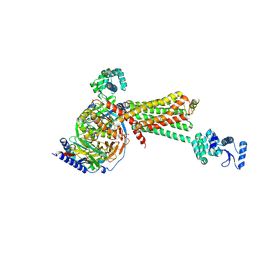 | | Crystal structure of the beta2 adrenergic receptor-Gs protein complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid antibody VHH fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Rasmussen, S.G.F, DeVree, B.T, Zou, Y, Kruse, A.C, Chung, K.Y, Kobilka, T.S, Thian, F.S, Chae, P.S, Pardon, E, Calinski, D, Mathiesen, J.M, Shah, S.T.A, Lyons, J.A, Caffrey, M, Gellman, S.H, Steyaert, J, Skiniotis, G, Weis, W.I, Sunahara, R.K, Kobilka, B.K. | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the beta2 adrenergic receptor-Gs protein complex
Nature, 477, 2011
|
|
3D23
 
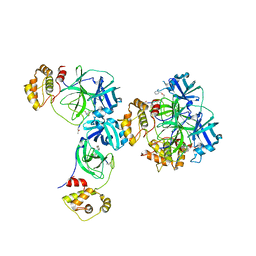 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
3KJ6
 
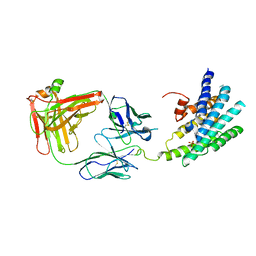 | | Crystal structure of a Methylated beta2 Adrenergic Receptor-Fab complex | | Descriptor: | Beta-2 adrenergic receptor, Fab heavy chain, Fab light chain, ... | | Authors: | Bokoch, M.P, Zou, Y, Rasmussen, S.G.F, Liu, C.W, Nygaard, R, Rosenbaum, D.M, Fung, J.J, Choi, H.-J, Thian, F.S, Kobilka, T.S, Puglisi, J.D, Weis, W.I, Pardo, L, Prosser, S, Mueller, L, Kobilka, B.K. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor.
Nature, 463, 2010
|
|
7VXY
 
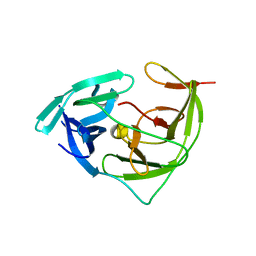 | | Zika virus NS2B/NS3 protease bZipro(C143S) in complex with D-RKOR | | Descriptor: | Peptide inhibitor, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Xiong, Y.C, Cheng, F, Zhang, J.Y, Su, H.X, Hu, H.C, Zou, Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure-based design of a novel inhibitor of the ZIKA virus NS2B/NS3 protease.
Bioorg.Chem., 128, 2022
|
|
7VXX
 
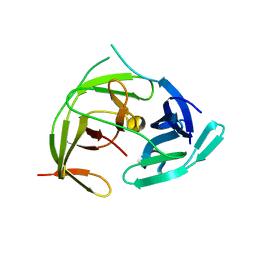 | | Zika virus NS2B/NS3 protease bZipro(C143S) in complex with 4-amino benzamidine | | Descriptor: | P-AMINO BENZAMIDINE, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Xiong, Y.C, Cheng, F, Zhang, J.Y, Su, H.X, Hu, H.C, Zou, Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design of a novel inhibitor of the ZIKA virus NS2B/NS3 protease.
Bioorg.Chem., 128, 2022
|
|
6N1T
 
 | | Toxoplasma gondii TS-DHFR in complex with selective inhibitor 3 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-{4-[3-(2-methoxypyrimidin-5-yl)phenyl]piperazin-1-yl}pyrimidine-2,4-diamine, ... | | Authors: | Hopper, A.T, Brockman, A, Wise, A, Gould, J, Barks, J, Radke, J.B, Sibley, L.D, Zou, Y, Thomas, S.B. | | Deposit date: | 2018-11-11 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of Selective Toxoplasma gondii Dihydrofolate Reductase Inhibitors for the Treatment of Toxoplasmosis.
J. Med. Chem., 62, 2019
|
|
6MXT
 
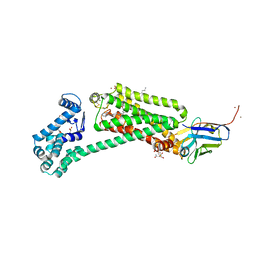 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
