1DG5
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND TRIMETHOPRIM | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1DG8
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1DF7
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND METHOTREXATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, METHOTREXATE, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-17 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1DG7
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND 4-BROMO WR99210 | | Descriptor: | 1-[3-(4-BROMO-PHENOXY)-PROPOXY]-6,6-DIMETHYL-1.6-DIHYDRO-[1,3,5]TRIAZINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1A2V
 
 | |
1P4X
 
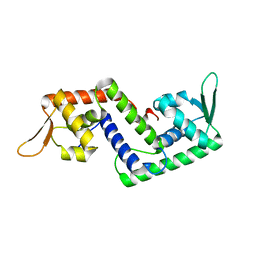 | | Crystal structure of SarS protein from Staphylococcus Aureus | | Descriptor: | staphylococcal accessory regulator A homologue | | Authors: | Li, R, Manna, A.C, Dai, S, Cheung, A.L, Zhang, G. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SarS protein from Staphylococcus aureus
J.BACTERIOL., 185, 2003
|
|
1QRD
 
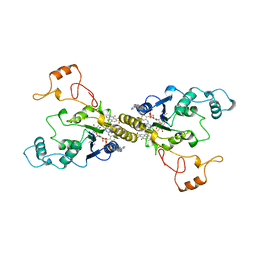 | | QUINONE REDUCTASE/FAD/CIBACRON BLUE/DUROQUINONE COMPLEX | | Descriptor: | CIBACRON BLUE, DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, R, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1995-07-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
4LTZ
 
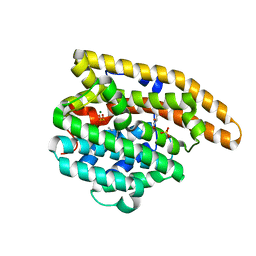 | | F95M Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LUU
 
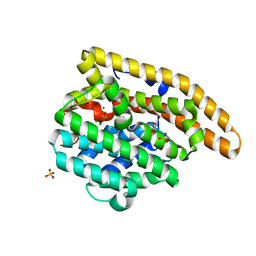 | | V329A Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LXW
 
 | | L72V Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-30 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LZ3
 
 | | F95H Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, HYDROGENPHOSPHATE ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LTV
 
 | | Crystal structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2) | | Descriptor: | Epi-isozizaene synthase, SULFATE ION | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LZC
 
 | | W325F Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LZ0
 
 | | A236G Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
7WCD
 
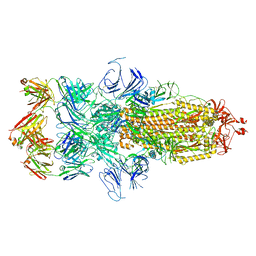 | | Cryo EM structure of SARS-CoV-2 spike in complex with TAU-2212 mAbs in conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Xiang, Y, Ma, B, Li, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
7WC0
 
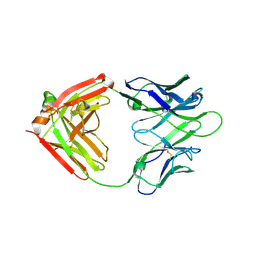 | |
6ITE
 
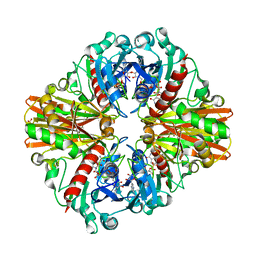 | | Crystal structure of group A Streptococcal surface dehydrogenase (SDH) | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Yuan, C, Li, R, Huang, M.D. | | Deposit date: | 2018-11-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structural determination of group A Streptococcal surface dehydrogenase and characterization of its interaction with urokinase-type plasminogen activator receptor.
Biochem.Biophys.Res.Commun., 510, 2019
|
|
6LSH
 
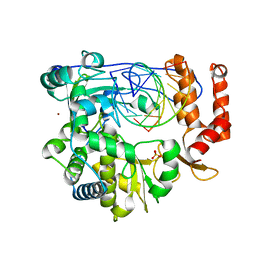 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6M form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Li, R, Wang, M, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
6LSG
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C0S6M form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-3'), ... | | Authors: | Li, R, Wang, M, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
5KZN
 
 | | Metabotropic Glutamate Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
1JV9
 
 | | NMR Structure of BPTI Mutant G37A | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Battiste, J.L, Li, R, Woodward, C. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-12 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A highly destabilizing mutation, G37A, of the bovine pancreatic trypsin inhibitor retains the average native conformation but greatly increases local flexibility
Biochemistry, 41, 2002
|
|
1JV8
 
 | | NMR Structure of BPTI Mutant G37A | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Battiste, J.L, Li, R, Woodward, C. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-12 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A highly destabilizing mutation, G37A, of the bovine pancreatic trypsin inhibitor retains the average native conformation but greatly increases local flexibility
Biochemistry, 41, 2002
|
|
1EKM
 
 | | CRYSTAL STRUCTURE AT 2.5 A RESOLUTION OF ZINC-SUBSTITUTED COPPER AMINE OXIDASE OF HANSENULA POLYMORPHA EXPRESSED IN ESCHERICHIA COLI | | Descriptor: | COPPER AMINE OXIDASE, ZINC ION | | Authors: | Chen, Z, Schwartz, B, Williams, N.K, Li, R, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2000-03-09 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure at 2.5 A resolution of zinc-substituted copper amine oxidase of Hansenula polymorpha expressed in Escherichia coli.
Biochemistry, 39, 2000
|
|
4YR6
 
 | | Fab fragment of 5G6 in complex with epitope peptide | | Descriptor: | ACE-LYS-LEU-ARG-GLY-VAL-LEU-GLN-GLY-HIS-LEU, GLYCEROL, heavy chain of 5G6, ... | | Authors: | Tao, Y, Mo, X, Li, R. | | Deposit date: | 2015-03-14 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for the specific inhibition of glycoprotein Ib alpha shedding by an inhibitory antibody.
Sci Rep, 6, 2016
|
|
