6YSF
 
 | |
6YSL
 
 | |
7B2A
 
 | |
7B29
 
 | |
7B2D
 
 | |
7B26
 
 | | CirpA1 in complex with pseudo-monomeric Properdin lacking TSR2-3 | | Descriptor: | CirpA1, Properdin, alpha-D-mannopyranose, ... | | Authors: | Lea, S.M, Johnson, S, Braunger, K. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of a family of tick-derived complement inhibitors targeting properdin.
Nat Commun, 13, 2022
|
|
7B28
 
 | |
6SD6
 
 | | Structure of VapBC from Shigella sonnei | | Descriptor: | Antitoxin, tRNA(fMet)-specific endonuclease VapC | | Authors: | Lea, S.M, Hollingshead, S. | | Deposit date: | 2019-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Polymorphisms in the VapBC toxin:antitoxin system mediate high frequency plasmid loss in Shigella sonnei
To Be Published
|
|
8FY3
 
 | | Structure of NOT1:NOT10:NOT11 module of the human CCR4-NOT complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Lea, S.M, Deme, J.C, Raisch, T, Pekovic, F, Valkov, E. | | Deposit date: | 2023-01-25 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure and assembly of the NOT10:11 module of the CCR4-NOT complex.
Commun Biol, 6, 2023
|
|
8FY4
 
 | | Structure of NOT1:NOT10:NOT11 module of the chicken CCR4-NOT complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Lea, S.M, Deme, J.C, Raisch, T, Levdansky, Y, Valkov, E. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structure and assembly of the NOT10:11 module of the CCR4-NOT complex.
Commun Biol, 6, 2023
|
|
1H2Q
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Powell, R, Spiller, O.B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H03
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H04
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, NICKEL (II) ION | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H2P
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
7YXX
 
 | |
7YXY
 
 | |
5NQX
 
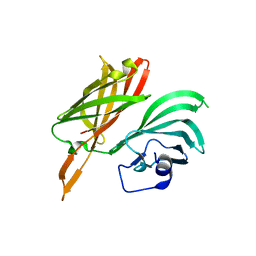 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 294. | | Descriptor: | Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein | | Authors: | Johnson, S, Jongerius, I, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
5NQP
 
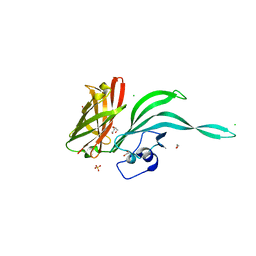 | | Structure of a fHbp(V1.4):PorA(P1.16) chimera. Fusion at fHbp position 151. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Factor H binding protein variant B16_001,Major outer membrane protein P.IA,Factor H binding protein variant B16_001, ... | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
1H8T
 
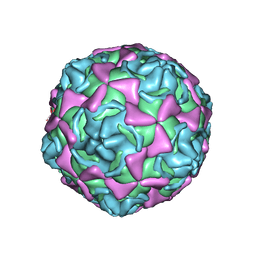 | | Echovirus 11 | | Descriptor: | 12-AMINO-DODECANOIC ACID, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Stuart, A, McKee, T, Williams, P.A, Harley, C, Stuart, D.I, Brown, T.D.K, Lea, S.M. | | Deposit date: | 2001-02-15 | | Release date: | 2002-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Determination of the Structure of a Decay Accelerating Factor-Binding Clinical Isolate of Echovirus 11 Allows Mapping of Mutants with Altered Receptor Requirements for Infection
J.Virol., 76, 2002
|
|
8SA4
 
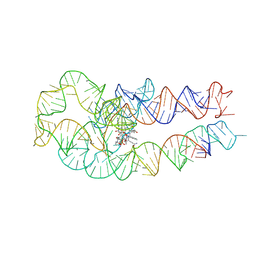 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
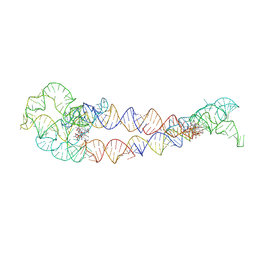 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA3
 
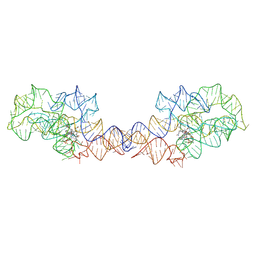 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
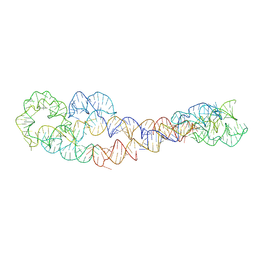 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA2
 
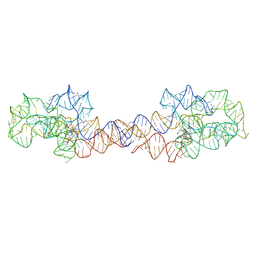 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8UCS
 
 | |
