3EG5
 
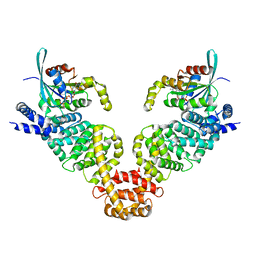 | | Crystal structure of MDIA1-TSH GBD-FH3 in complex with CDC42-GMPPNP | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lammers, M, Meyer, S, Kuehlmann, D, Wittinghofer, A. | | Deposit date: | 2008-09-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity of Interactions between mDia Isoforms and Rho Proteins
J.Biol.Chem., 283, 2008
|
|
2BAP
 
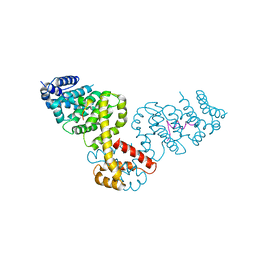 | |
2X2A
 
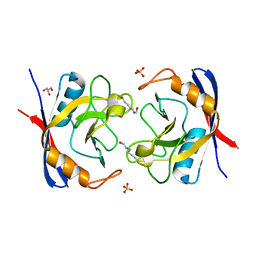 | | Free acetyl-CypA trigonal form | | Descriptor: | GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A, SULFATE ION | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerization.
Nat.Chem.Biol., 6, 2010
|
|
2X2D
 
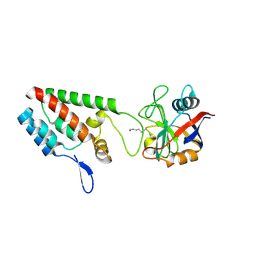 | | acetyl-CypA:HIV-1 N-term capsid domain complex | | Descriptor: | CAPSID PROTEIN P24, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerisation
Nat.Chem.Biol., 6, 2010
|
|
2X2C
 
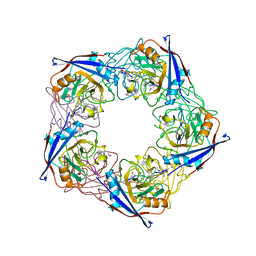 | | acetyl-CypA:cyclosporine complex | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerisation
Nat.Chem.Biol., 6, 2010
|
|
2X25
 
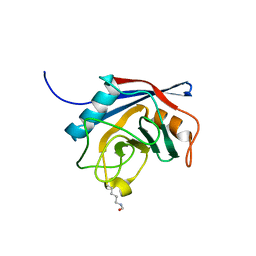 | | Free acetyl-CypA orthorhombic form | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-11 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerization.
Nat.Chem.Biol., 6, 2010
|
|
4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
1Z2C
 
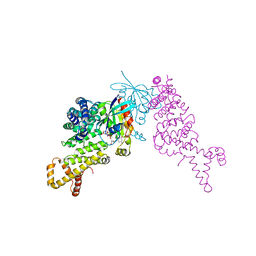 | | Crystal structure of mDIA1 GBD-FH3 in complex with RhoC-GMPPNP | | Descriptor: | Diaphanous protein homolog 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Rose, R, Weyand, M, Lammers, M, Ishizaki, T, Ahmadian, M.R, Wittinghofer, A. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and mechanistic insights into the interaction between Rho and mammalian Dia.
Nature, 435, 2005
|
|
5FR2
 
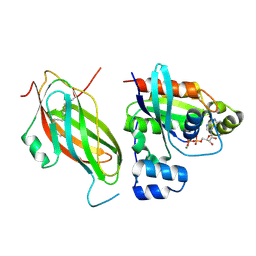 | | Farnesylated RhoA-GDP in complex with RhoGDI-alpha, lysine acetylated at K178 | | Descriptor: | FARNESYL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kuhlmann, N, Wroblowski, S, Knyphausen, P, de Boor, S, Brenig, J, Zienert, A.Y, Meyer-Teschendorf, K, Praefcke, G.J.K, Nolte, H, Krueger, M, Schacherl, M, Baumann, U, James, L.C, Chin, J.W, Lammers, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural and Mechanistic Insights Into the Regulation of the Fundamental Rho-Regulator Rhogdi Alpha by Lysine Acetylation.
J.Biol.Chem., 291, 2016
|
|
5FYQ
 
 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
7QE9
 
 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 N34S (SPINK1 N34S) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
7QE8
 
 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 (SPINK1) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
7P97
 
 | | Structure of 3-phospho-D-glycerate guanylyltransferase with product 3-GPPG bound | | Descriptor: | 3-(guanosine-5'-diphospho)-D-glycerate, 3-phospho-D-glycerate guanylyltransferase, CHLORIDE ION, ... | | Authors: | Palm, G.J, Berndt, L, Lammers, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Diversification by CofC and Control by CofD Govern Biosynthesis and Evolution of Coenzyme F 420 and Its Derivative 3PG-F 420.
Mbio, 13, 2022
|
|
7AC0
 
 | | Epoxide hydrolase CorEH without ligand | | Descriptor: | Soluble epoxide hydrolase | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Promiscuous Dehalogenase Activity of the Epoxide Hydrolase CorEH from Corynebacterium sp. C12
Acs Catalysis, 11, 2021
|
|
6Y9K
 
 | |
5FR1
 
 | | Double acetylated RhoGDI-alpha in complex with RhoA-GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RHO GDP-DISSOCIATION INHIBITOR 1, ... | | Authors: | Kuhlmann, N, Wroblowski, S, Lammers, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rhogdi Alpha Acetylation at K127 and K141 Affects Binding Towards Non-Prenylated Rhoa.
Biochemistry, 55, 2016
|
|
7ATL
 
 | | EstCE1, a hydrolase with promiscuous acyltransferase activity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Esterase, ... | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | Discovery and Design of Family VIII Carboxylesterases as Highly Efficient Acyltransferases.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7Z9F
 
 | | Human anionic trypsin after autoproteolysis at Arg122 | | Descriptor: | CALCIUM ION, Trypsin-2 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2022-03-21 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of the Pancreatitis-Associated Autoproteolytic Failsafe Mechanism in Human Anionic Trypsin.
J Inflamm Res, 15, 2022
|
|
2WLW
 
 | | Structure of the N-terminal capsid domain of HIV-2 | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Price, A.J, Marzetta, F, Lammers, M, Ylinen, L.M.J, Schaller, T, Wilson, S.J, Towers, G.J, James, L.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active Site Remodelling Switches HIV Specificity of Antiretroviral Trimcyp
Nat.Struct.Mol.Biol., 16, 2009
|
|
2WLV
 
 | | Structure of the N-terminal capsid domain of HIV-2 | | Descriptor: | GAG POLYPROTEIN | | Authors: | Price, A.J, Marzetta, F, Lammers, M, Ylinen, L.M.J, Schaller, T, Wilson, S.J, Towers, G.J, James, L.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-22 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Active Site Remodelling Switches HIV Specificity of Antiretroviral Trimcyp
Nat.Struct.Mol.Biol., 16, 2009
|
|
7CUV
 
 | |
7E31
 
 | | Crystal structure of a novel alpha/beta hydrolase mutant in apo form | | Descriptor: | TRIETHYLENE GLYCOL, alpha/beta hydrolase | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7E30
 
 | | Crystal structure of a novel alpha/beta hydrolase in apo form in complex with citrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, SULFATE ION, ... | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
6Z1B
 
 | |
7W1K
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca | | Descriptor: | Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
