2RRE
 
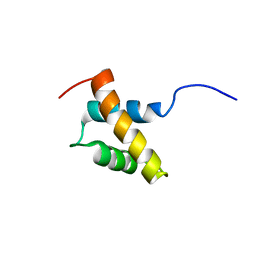 | | Structure and function of the N-terminal nucleolin binding domain of nuclear valocine containing protein like 2 (NVL2) harboring a nucleolar localization signal | | Descriptor: | Putative uncharacterized protein | | Authors: | Fujiwara, Y, Fujiwara, K, Goda, N, Iwaya, N, Tenno, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2010-08-03 | | Release date: | 2011-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal nucleolin binding domain of nuclear valosin-containing protein-like 2 (NVL2) harboring a nucleolar localization signal
J.Biol.Chem., 286, 2011
|
|
3E7K
 
 | |
3AXA
 
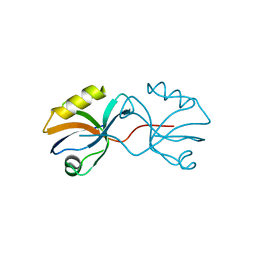 | | Crystal structure of afadin PDZ domain in complex with the C-terminal peptide from nectin-3 | | Descriptor: | Afadin, Nectin-3 | | Authors: | Fujiwara, Y, Goda, N, Narita, H, Satomura, K, Nakagawa, A, Sakisaka, T, Suzuki, M, Hiroaki, H. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of afadin PDZ domain-nectin-3 complex shows the structural plasticity of the ligand-binding site.
Protein Sci., 24, 2015
|
|
3VMX
 
 | | Crystal Structure of a parallel coiled-coil dimerization domain from the voltage-gated proton channel | | Descriptor: | Voltage-gated hydrogen channel 1 | | Authors: | Fujiwara, Y, Takeshita, K, Kobayashi, M, Okamura, Y, Nakagawa, A. | | Deposit date: | 2011-12-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The cytoplasmic coiled-coil mediates cooperative gating temperature sensitivity in the voltage-gated H(+) channel Hv1
Nat Commun, 3, 2012
|
|
3VMZ
 
 | |
3VMY
 
 | |
3VN0
 
 | |
3VYI
 
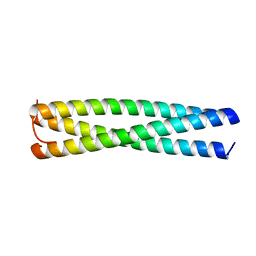 | |
3DVJ
 
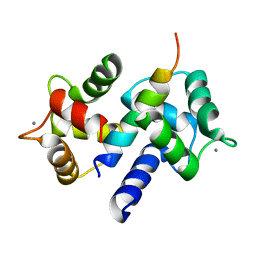 | | Crystal Structure of Ca2+/CaM-CaV2.2 IQ domain (without cloning artifact, HM to TV) complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent N-type calcium channel subunit alpha-1B | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVE
 
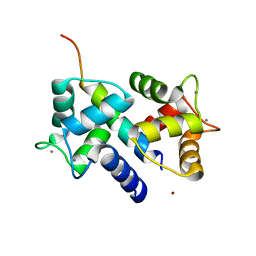 | | Crystal Structure of Ca2+/CaM-CaV2.2 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, NICKEL (II) ION, ... | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVM
 
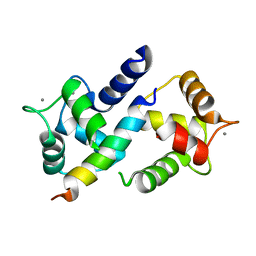 | | Crystal Structure of Ca2+/CaM-CaV2.1 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent P/Q-type calcium channel subunit alpha-1A | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVK
 
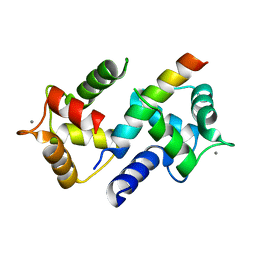 | | Crystal Structure of Ca2+/CaM-CaV2.3 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent R-type calcium channel subunit alpha-1E | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
4Z2M
 
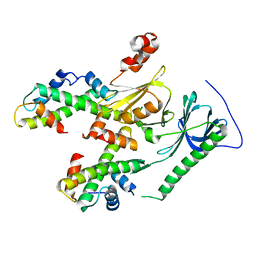 | | Crystal structure of human SPT16 Mid-AID/H3-H4 tetramer FACT Histone complex | | Descriptor: | FACT complex subunit SPT16, Histone H3.1, Histone H4 | | Authors: | Tsunaka, Y, Fujiwara, Y, Oyama, T, Hirose, S, Morikawa, K. | | Deposit date: | 2015-03-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Integrated molecular mechanism directing nucleosome reorganization by human FACT.
Genes Dev., 30, 2016
|
|
4Z2N
 
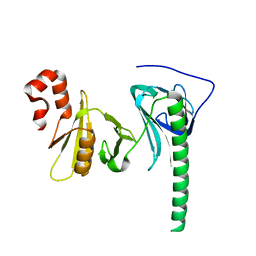 | | Crystal structure of human FACT SPT16 middle domain | | Descriptor: | FACT complex subunit SPT16 | | Authors: | Tsunaka, Y, Fujiwara, Y, Oyama, T, Hirose, S, Morikawa, K. | | Deposit date: | 2015-03-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Integrated molecular mechanism directing nucleosome reorganization by human FACT.
Genes Dev., 30, 2016
|
|
1IXS
 
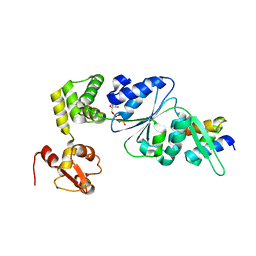 | | Structure of RuvB complexed with RuvA domain III | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1IXR
 
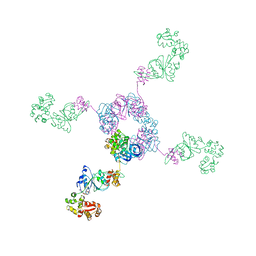 | | RuvA-RuvB complex | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
3WKV
 
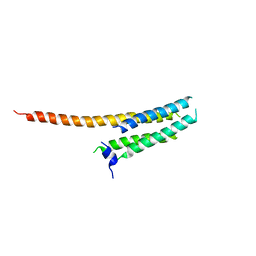 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | Descriptor: | Ion channel | | Authors: | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.453 Å) | | Cite: | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7PL9
 
 | |
2RPA
 
 | | The solution structure of N-terminal domain of microtubule severing enzyme | | Descriptor: | Katanin p60 ATPase-containing subunit A1 | | Authors: | Iwaya, N, Kuwahara, Y, Unzai, S, Nagata, T, Tomii, K, Goda, N, Tochio, H, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2008-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A common substrate recognition mode conserved between katanin P60 and VPS4 governs microtubule severing and membrane skeleton reorganization
J.Biol.Chem., 285, 2010
|
|
5WIR
 
 | | Structure of the TRF1-TERB1 interface | | Descriptor: | TERB1-TBM, Telomeric repeat-binding factor 1 | | Authors: | Nandakumar, J, Pendlebury, D.F, Smith, E.M, Tesmer, V.M. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dissecting the telomere-inner nuclear membrane interface formed in meiosis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5H36
 
 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Rhodobacter sphaeroides | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Uncharacterized protein TRIC | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
5H35
 
 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Sulfolobus solfataricus | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
5WZY
 
 | | Crystal structure of the P2X4 receptor from zebrafish in the presence of CTP at 2.8 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural insights into the nucleotide base specificity of P2X receptors
Sci Rep, 7, 2017
|
|
5F1C
 
 | | Crystal structure of an invertebrate P2X receptor from the Gulf Coast tick in the presence of ATP and Zn2+ ion at 2.9 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein, ... | | Authors: | Kasuya, G, Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into Divalent Cation Modulations of ATP-Gated P2X Receptor Channels
Cell Rep, 14, 2016
|
|
2VNS
 
 | | Crystal Structure of the Membrane Proximal Oxidoreductase Domain of Human Steap3, the Dominant Ferric Reductase of the Erythroid Transferrin Cycle | | Descriptor: | CITRIC ACID, METALLOREDUCTASE STEAP3 | | Authors: | Sendamarai, A.K, Ohgami, R.S, Fleming, M.D, Lawrence, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Membrane Proximal Oxidoreductase Domain of Human Steap3, the Dominant Ferrireductase of the Erythroid Transferrin Cycle
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
