1XV2
 
 | | Crystal Structure of a Protein of Unknown Function Similar to Alpha-acetolactate Decarboxylase from Staphylococcus aureus | | Descriptor: | ZINC ION, hypothetical protein, similar to alpha-acetolactate decarboxylase | | Authors: | Duke, N.E.C, Zhang, R, Quartey, P, Collart, F, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a hypothetical protein similar to alpha-acetolactate
To be Published
|
|
2ZC2
 
 | | Crystal structure of DnaD-like replication protein from Streptococcus mutans UA159, gi 24377835, residues 127-199 | | Descriptor: | DnaD-like replication protein, ZINC ION | | Authors: | Duke, N.E.C, Clancy, S, Duggan, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of DnaD-like replication protein from Streptococcus mutans UA159.
To be Published
|
|
3FKF
 
 | |
2I9D
 
 | |
2I7R
 
 | |
1RWJ
 
 | | c7-type three-heme cytochrome domain | | Descriptor: | Cytochrome c family protein, HEME C | | Authors: | Pokkuluri, P.R, Londer, Y.Y, Duke, N.E.C, Erickson, J, Pessanha, M, Salgueiro, C.A, Schiffer, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-08-03 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a novel c7-type three-heme cytochrome domain from a multidomain cytochrome c polymer.
Protein Sci., 13, 2004
|
|
3SOZ
 
 | | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2 | | Descriptor: | Cytoplasmic Protein STM1381, GLYCEROL | | Authors: | Joachimiak, A, Duke, N.E.C, Jedrzejczak, R, Li, H, Adkins, J, Brown, R, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2
To be Published
|
|
3OT6
 
 | | Crystal Structure of an enoyl-CoA hydratase/isomerase family protein from Psudomonas syringae | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Joachimiak, A, Duke, N.E.C, Stein, A, Chhor, G, Freeman, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-09-10 | | Release date: | 2010-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an enoyl-CoA hydratase/isomerase family protein from Psudomonas syringae
To be Published
|
|
3OOV
 
 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
3OVK
 
 | |
3PN9
 
 | |
3R0A
 
 | |
3IDD
 
 | |
3KD8
 
 | |
3LM7
 
 | | Crystal Structure of DUF1341 representative, from Yersinia enterocolitica subsp. enterocolitica 8081 | | Descriptor: | BROMIDE ION, POTASSIUM ION, putative 4-Hydroxy-2-oxoglutarate aldolase / 2-dehydro-3-deoxyphosphogluconate aldolase | | Authors: | Joachimiak, A, Duke, N.E.C, Feldmann, B, Wu, R, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of DUF1341 representative, from Yersinia enterocolitica subsp. enterocolitica 8081
To be Published
|
|
1YLX
 
 | |
4HTQ
 
 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 6.70 x 10e+11 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.Z, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HTN
 
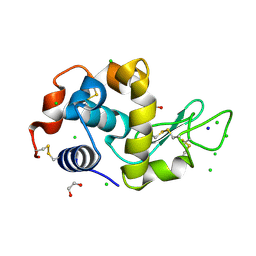 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 1.32 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HTK
 
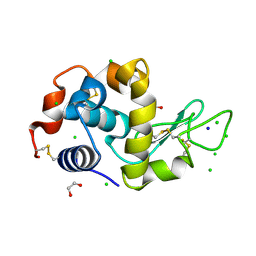 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 2.17 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1OS6
 
 | | Cytochrome c7 (PpcA) from Geobacter sulfurreducens | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, HEME C, PpcA, ... | | Authors: | Pokkuluri, P.R, Londer, Y.Y, Duke, N.E.C, Long, W.C, Schiffer, M. | | Deposit date: | 2003-03-18 | | Release date: | 2004-02-03 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Family of cytochrome c7-type proteins from Geobacter sulfurreducens: structure of one cytochrome c7 at 1.45 A resolution.
Biochemistry, 43, 2004
|
|
6NEX
 
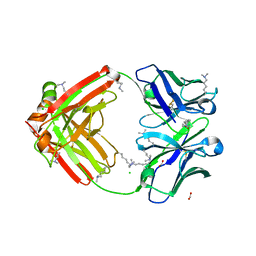 | | Fab fragment of anti-cocaine antibody h2E2 | | Descriptor: | ACETATE ION, Anitgen binding fragment light chain, Antigen binding fragment heavy chain, ... | | Authors: | Pokkuluri, P.R, Tan, K. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NFN
 
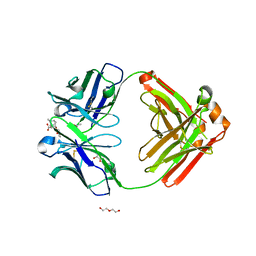 | | Fab fragment of anti-cocaine antibody h2E2 bound to benzoylecgonine | | Descriptor: | 3-(BENZOYLOXY)-8-METHYL-8-AZABICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pokkuluri, P.R, Tan, K. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6U97
 
 | | Structure of OmcF_H47I mutant | | Descriptor: | Lipoprotein cytochrome c, 1 heme-binding site, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pokkuluri, P.R. | | Deposit date: | 2019-09-06 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Modulation of the Redox Potential and Electron/Proton Transfer Mechanisms in the Outer Membrane Cytochrome OmcF FromGeobacter sulfurreducens.
Front Microbiol, 10, 2019
|
|
